3FMP
 
 | | Crystal structure of the nucleoporin Nup214 in complex with the DEAD-box helicase Ddx19 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DDX19B, Nuclear pore complex protein Nup214 | | Authors: | Napetschnig, J, Debler, E.W, Blobel, G, Hoelz, A. | | Deposit date: | 2008-12-22 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structural and functional analysis of the interaction between the nucleoporin Nup214 and the DEAD-box helicase Ddx19.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6BZ2
 
 | | Crystal structure of wild-type HIV-1 protease with a novel HIV-1 inhibitor GRL-14213A of 6-5-5-ring fused crown-like tetrahydropyranofuran as the P2-ligand, a cyclopropylaminobenzothiazole as the P2'-ligand and 3,5-difluorophenylmethyl as the P1-ligand | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2017-12-22 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Design of Highly Potent, Dual-Acting and Central-Nervous-System-Penetrating HIV-1 Protease Inhibitors with Excellent Potency against Multidrug-Resistant HIV-1 Variants.
ChemMedChem, 13, 2018
|
|
3FNJ
 
 | | Crystal structure of the full-length lp_1913 protein from Lactobacillus plantarum, Northeast Structural Genomics Consortium Target LpR140 | | Descriptor: | protein lp_1913 | | Authors: | Seetharaman, J, Chen, Y, Forouhar, F, Sahdev, S, Janjua, H, Xiao, R, Ciccosanti, C, Foote, E.L, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-25 | | Release date: | 2009-01-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the full-length lp_1913 protein from Lactobacillus plantarum, Northeast Structural Genomics Consortium Target LpR140
To be Published
|
|
6GAY
 
 | |
3FEQ
 
 | | Crystal structure of uncharacterized protein eah89906 | | Descriptor: | PUTATIVE AMIDOHYDROLASE, ZINC ION | | Authors: | Patskovsky, Y, Bonanno, J, Romero, R, Freeman, J, Lau, C, Smith, D, Bain, K, Wasserman, S.R, Raushel, F, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-30 | | Release date: | 2008-12-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily .
Biochemistry, 49, 2010
|
|
6BW6
 
 | | Human GPT (DPAGT1) H129 variant in complex with tunicamycin | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Tunicamycin, UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase | | Authors: | Yoo, J, Kuk, A.C.Y, Mashalidis, E.H, Lee, S.-Y. | | Deposit date: | 2017-12-14 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | GlcNAc-1-P-transferase-tunicamycin complex structure reveals basis for inhibition of N-glycosylation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6GB3
 
 | |
3FH6
 
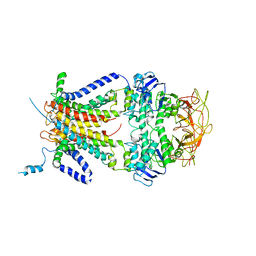 | | Crystal structure of the resting state maltose transporter from E. coli | | Descriptor: | Maltose transport system permease protein malF, Maltose transport system permease protein malG, Maltose/maltodextrin import ATP-binding protein malK | | Authors: | Khare, D, Oldham, M.L, Orelle, C, Davidson, A.L, Chen, J. | | Deposit date: | 2008-12-08 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Alternating access in maltose transporter mediated by rigid-body rotations.
Mol.Cell, 33, 2009
|
|
3F4D
 
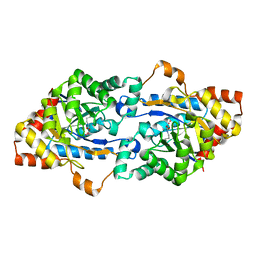 | | Crystal structure of organophosphorus hydrolase from Geobacillus stearothermophilus strain 10 | | Descriptor: | COBALT (II) ION, Organophosphorus hydrolase | | Authors: | Hawwa, R, Aikens, J, Turner, R.J, Santarsiero, B, Mesecar, A. | | Deposit date: | 2008-10-31 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural basis for thermostability revealed through the identification and characterization of a highly thermostable phosphotriesterase-like lactonase from Geobacillus stearothermophilus.
Arch.Biochem.Biophys., 488, 2009
|
|
6BXB
 
 | | Crystal structure of an extended b3 integrin P33 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Chimera protein of Integrin beta-3 and Integrin alpha-L, ... | | Authors: | Zhou, D, Zhu, J. | | Deposit date: | 2017-12-18 | | Release date: | 2018-08-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of an extended beta3integrin.
Blood, 132, 2018
|
|
3F6M
 
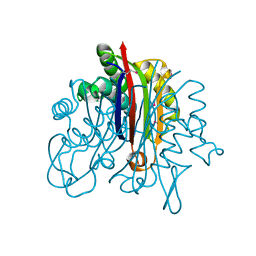 | | Crystal Structure of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase IspF from Yersinia pestis | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase | | Authors: | Kim, Y, Maltseva, N, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-11-06 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Crystal Structure of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase IspF from Yersinia pestis
To be Published
|
|
6BY7
 
 | | Folding DNA into a lipid-conjugated nano-barrel for controlled reconstitution of membrane proteins | | Descriptor: | DNA (26-MER), DNA (27-MER), DNA (29-MER), ... | | Authors: | Dong, Y, Chen, S, Zhang, S, Sodroski, J, Yang, Z, Liu, D, Mao, Y. | | Deposit date: | 2017-12-20 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Folding DNA into a Lipid-Conjugated Nanobarrel for Controlled Reconstitution of Membrane Proteins.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
3FJD
 
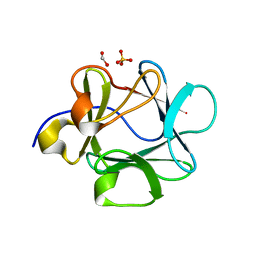 | |
3FJJ
 
 | |
6C5R
 
 | | Crystal structure of the soluble domain of the mitochondrial calcium uniporter | | Descriptor: | calcium uniporter | | Authors: | Fan, C, Fan, M, Fastman, N, Zhang, J, Feng, L. | | Deposit date: | 2018-01-16 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.09608316 Å) | | Cite: | X-ray and cryo-EM structures of the mitochondrial calcium uniporter.
Nature, 559, 2018
|
|
3FLE
 
 | | SE_1780 protein of unknown function from Staphylococcus epidermidis. | | Descriptor: | SE_1780 protein | | Authors: | Osipiuk, J, Hatzos, C, Clancy, S, Kim, Y, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-18 | | Release date: | 2009-01-13 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | X-ray crystal structure of SE_1780 protein of unknown function from Staphylococcus epidermidis.
To be Published
|
|
6G0D
 
 | |
6G0F
 
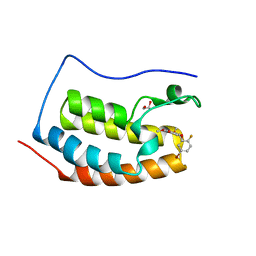 | | BRD4 (BD1) in complex with docking-derived ligand | | Descriptor: | (4~{R})-4-(5-bromanyl-2-fluoranyl-phenyl)-5,6,7-trimethoxy-3,4-dihydro-1~{H}-quinolin-2-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Pretzel, J, Humbeck, L. | | Deposit date: | 2018-03-18 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.618 Å) | | Cite: | Discovery of an Unexpected Similarity in Ligand Binding Between BRD4 and PPARgamma
Chemrxiv, 2019
|
|
6FNE
 
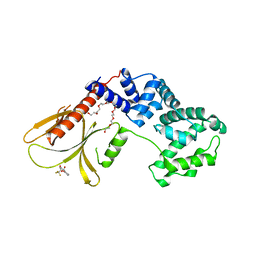 | | Structure of human Brag2 (Sec7-PH domains) with the inhibitor Bragsin bound to the PH domain | | Descriptor: | (2~{S})-6-methyl-5-nitro-2-(trifluoromethyl)-2,3-dihydrochromen-4-one, IQ motif and SEC7 domain-containing protein 1, NONAETHYLENE GLYCOL | | Authors: | Nawrotek, A, Zeghouf, M, Cherfils, J. | | Deposit date: | 2018-02-03 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | PH-domain-binding inhibitors of nucleotide exchange factor BRAG2 disrupt Arf GTPase signaling.
Nat.Chem.Biol., 15, 2019
|
|
6C1Y
 
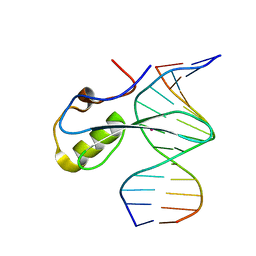 | | mbd of human mecp2 in complex with methylated DNA | | Descriptor: | 12-mer DNA, Methyl-CpG-binding protein 2, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Bian, C, Tempel, W, Wernimont, A.K, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-01-05 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Plasticity at the DNA recognition site of the MeCP2 mCG-binding domain.
Biochim Biophys Acta Gene Regul Mech, 1862, 2019
|
|
3FFO
 
 | | F17b-G lectin domain with bound GlcNAc(beta1-2)man | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose, Adhesin, ... | | Authors: | Buts, L, De Boer, A, Olsson, J.D.M, Jonckheere, W, De Kerpel, M, De Genst, E, Guerardel, Y, Willaert, R, Wyns, L, Wuhrer, M, Oscarson, S, De Greve, H, Bouckaert, J. | | Deposit date: | 2008-12-04 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Sampling of Glycan Interaction Profiles Reveals Mucosal Receptors for Fimbrial Adhesins of Enterotoxigenic Escherichia coli.
Biology (Basel), 2, 2013
|
|
3FGM
 
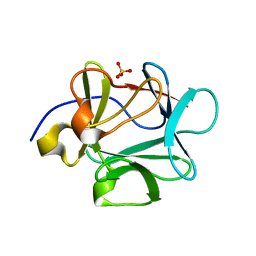 | |
6C3P
 
 | | Cryo-EM structure of human KATP bound to ATP and ADP in propeller form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Lee, K.P.K, Chen, J, MacKinnon, R. | | Deposit date: | 2018-01-10 | | Release date: | 2018-01-24 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Molecular structure of human KATP in complex with ATP and ADP.
Elife, 6, 2017
|
|
3FH3
 
 | | Crystal structure of a putative ECF-type sigma factor negative effector from Bacillus anthracis str. Sterne | | Descriptor: | NICKEL (II) ION, putative ECF-type sigma factor negative effector | | Authors: | Nocek, B, Kim, Y, Joachimiak, G, Du, J, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-08 | | Release date: | 2009-01-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Crystal structure of a putative ECF-type sigma factor negative effector from Bacillus anthracis str. Sterne
To be Published
|
|
3F4K
 
 | | Crystal structure of a probable methyltransferase from Bacteroides thetaiotaomicron. Northeast Structural Genomics target BtR309. | | Descriptor: | Putative methyltransferase | | Authors: | Seetharaman, J, Lew, S, Wang, H, Janjua, H, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-31 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a probable methyltransferase from Bacteroides thetaiotaomicron. Northeast Structural Genomics target BtR309.
To be Published
|
|
