3VCZ
 
 | | 1.80 Angstrom resolution crystal structure of a putative translation initiation inhibitor from Vibrio vulnificus CMCP6 | | Descriptor: | CALCIUM ION, Endoribonuclease L-PSP, GLYCEROL, ... | | Authors: | Halavaty, A.S, Minasov, G, Filippova, E.V, Dubrovska, I, Winsor, J, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-04 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.80 Angstrom resolution crystal structure of a putative translation initiation inhibitor from Vibrio vulnificus CMCP6
To be Published
|
|
3VGP
 
 | | Crystal structure of the C-terminal globular domain of oligosaccharyltransferase (AF_0329) from Archaeoglobus fulgidus | | Descriptor: | Transmembrane oligosaccharyl transferase, putative | | Authors: | Matsumoto, S, Igura, M, Nyirenda, J, Yuzawa, S, Noda, N.N, Inagaki, F, Kohda, D. | | Deposit date: | 2011-08-18 | | Release date: | 2012-07-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the C-Terminal Globular Domain of Oligosaccharyltransferase from Archaeoglobus fulgidus at 1.75 A Resolution
Biochemistry, 51, 2012
|
|
1QWJ
 
 | | The Crystal Structure of Murine CMP-5-N-Acetylneuraminic Acid Synthetase | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE-5-N-ACETYLNEURAMINIC ACID, cytidine monophospho-N-acetylneuraminic acid synthetase | | Authors: | Krapp, S, Muenster-Kuehnel, A.K, Kaiser, J.T, Huber, R, Tiralongo, J, Gerardy-Schahn, R, Jacob, U. | | Deposit date: | 2003-09-02 | | Release date: | 2003-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of Murine CMP-5-N-acetylneuraminic Acid Synthetase
J.Mol.Biol., 334, 2003
|
|
3VI3
 
 | | Crystal structure of alpha5beta1 integrin headpiece (ligand-free form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Nagae, M, Nogi, T, Takagi, J. | | Deposit date: | 2011-09-21 | | Release date: | 2012-02-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of alpha5beta1 integrin ectodomain: Atomic details of the fibronectin receptor
J.Cell Biol., 197, 2012
|
|
3GMF
 
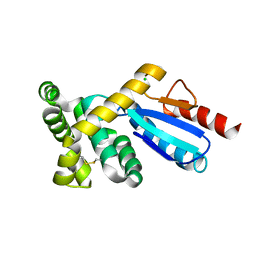 | | Crystal structure of protein-disulfide isomerase from Novosphingobium aromaticivorans | | Descriptor: | CHLORIDE ION, Protein-disulfide isomerase | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-13 | | Release date: | 2009-03-24 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of protein-disulfide isomerase from Novosphingobium aromaticivorans
To be Published
|
|
3GS1
 
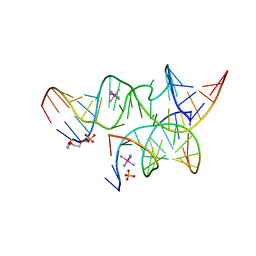 | | An all-RNA Hairpin Ribozyme with mutation A38N1dA | | Descriptor: | 2-[2-(2-HYDROXYETHOXY)ETHOXY]ETHYL DIHYDROGEN PHOSPHATE, COBALT HEXAMMINE(III), RNA (5'-R(*CP*GP*GP*UP*GP*AP*GP*AP*AP*GP*GP*G)-3'), ... | | Authors: | Spitale, R.C, Volpini, R, Heller, M.G, Krucinska, J, Cristalli, G, Wedekind, J.E. | | Deposit date: | 2009-03-26 | | Release date: | 2009-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Identification of an imino group indispensable for cleavage by a small ribozyme.
J.Am.Chem.Soc., 131, 2009
|
|
1R0C
 
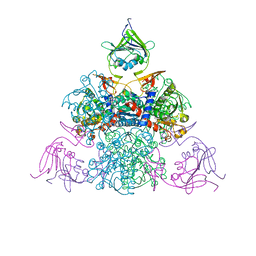 | |
3VKK
 
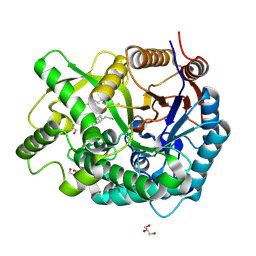 | | Crystal Structure Of The Covalent Intermediate Of Human Cytosolic Beta-Glucosidase-mannose complex | | Descriptor: | CHLORIDE ION, Cytosolic beta-glucosidase, GLYCEROL, ... | | Authors: | Noguchi, J, Hayashi, Y, Okino, N, Ito, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2011-11-17 | | Release date: | 2012-11-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for inhibition mechanism of human cytosolic beta-glucosidase by monnoside
To be Published
|
|
3GQQ
 
 | | Crystal structure of the human retinal protein 4 (unc-119 homolog A). Northeast Structural Genomics Consortium target HR3066a | | Descriptor: | Protein unc-119 homolog A, UNKNOWN LIGAND | | Authors: | Vorobiev, S.M, Chen, Y, Seetharaman, J, Shastry, R, Foote, E.L, Ciccosanti, C, Sahdev, S, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-03-24 | | Release date: | 2009-04-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.945 Å) | | Cite: | Crystal structure of the human retinal protein 4 (unc-119 homolog A).
To be Published
|
|
3GRO
 
 | | Human palmitoyl-protein thioesterase 1 | | Descriptor: | Palmitoyl-protein thioesterase 1, UNKNOWN ATOM OR ION | | Authors: | Dobrovetsky, E, Seitova, A, Tong, Y, Tempel, W, Dong, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Cossar, D, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-03-26 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Human palmitoyl-protein thioesterase 1
To be Published
|
|
3GTZ
 
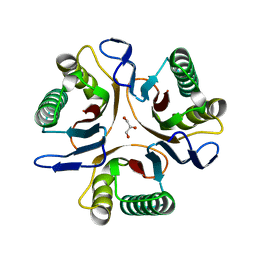 | | Crystal structure of a putative translation initiation inhibitor from Salmonella typhimurium | | Descriptor: | GLYCEROL, Putative translation initiation inhibitor | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Miller, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-28 | | Release date: | 2009-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a putative translation initiation inhibitor from Salmonella typhimurium
To be Published
|
|
3V7J
 
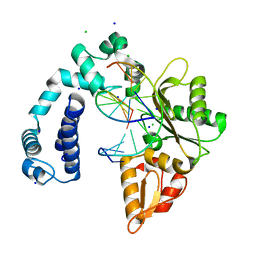 | | Co-crystal structure of Wild Type Rat polymerase beta: Enzyme-DNA binary complex | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*AP*TP*GP*TP*GP*AP*GP*T)-3'), DNA (5'-D(P*CP*AP*AP*AP*CP*TP*CP*AP*CP*AP*TP*A)-3'), ... | | Authors: | Rangarajan, S, Jaeger, J. | | Deposit date: | 2011-12-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallographic studies of K72E mutant DNA polymerase explain loss of lyase function and reveal changes in the overall conformational state of the polymerase domain
To be Published
|
|
3GU3
 
 | | Crystal Structure of the methyltransferase BC_2162 in complex with S-Adenosyl-L-Homocysteine from Bacillus cereus, Northeast Structural Genomics Consortium Target BcR20 | | Descriptor: | ACETATE ION, Methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Ciano, C, Ma, L, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-03-28 | | Release date: | 2009-04-07 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Northeast Structural Genomics Consortium Target BcR20
To be Published
|
|
1S6R
 
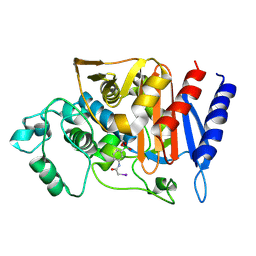 | | 908R class c beta-lactamase bound to iodo-acetamido-phenyl boronic acid | | Descriptor: | 4-IODO-ACETAMIDO PHENYLBORONIC ACID, beta-lactamase | | Authors: | Wouters, J. | | Deposit date: | 2004-01-27 | | Release date: | 2004-02-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal structure of Enterobacter cloacae 908R class C beta-lactamase bound to iodo-acetamido-phenyl boronic acid, a transition-state analogue.
Cell.Mol.Life Sci., 60, 2003
|
|
1S79
 
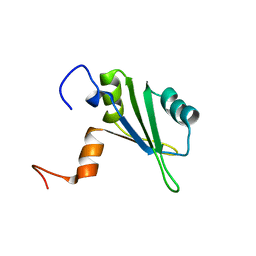 | | Solution structure of the central RRM of human La protein | | Descriptor: | Lupus La protein | | Authors: | Alfano, C, Sanfelice, D, Babon, J, Kelly, G, Jacks, A, Curry, S, Conte, M.R. | | Deposit date: | 2004-01-29 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein.
Nat.Struct.Mol.Biol., 11, 2004
|
|
4KMS
 
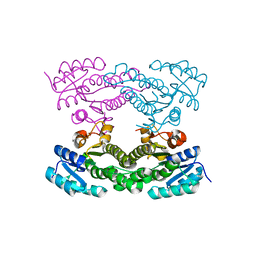 | | Crystal structure of Acetoacetyl-CoA reductase from Rickettsia felis | | Descriptor: | Acetoacetyl-CoA reductase | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID), Abendroth, J, Lukacs, C, Edwards, T.E, Lorimer, D. | | Deposit date: | 2013-05-08 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of acetoacetyl-CoA reductase from Rickettsia felis.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
3H00
 
 | |
3VJ6
 
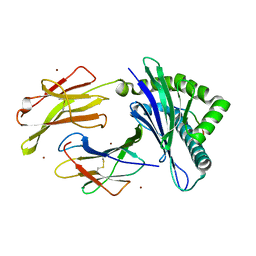 | | Structure of the MHC class Ib molecule Qa-1b | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-37 alpha chain, ... | | Authors: | Zeng, L, Clements, C.S, Rossjohn, J. | | Deposit date: | 2011-10-12 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A structural basis for antigen presentation by the MHC class Ib molecule, Qa-1b
J.Immunol., 188, 2012
|
|
3H1U
 
 | | Structure of ubiquitin in complex with Cd ions | | Descriptor: | CADMIUM ION, Ubiquitin | | Authors: | Qureshi, I.A, Ferron, F, Cheung, P, Lescar, J. | | Deposit date: | 2009-04-14 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic structure of ubiquitin in complex with cadmium ions
BMC RES NOTES, 2, 2009
|
|
3H2B
 
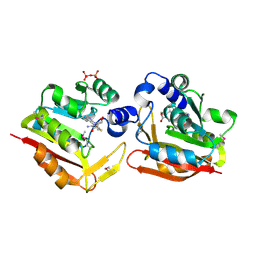 | | Crystal structure of the SAM-dependent methyltransferase cg3271 from Corynebacterium glutamicum in complex with S-adenosyl-L-homocysteine and pyrophosphate. Northeast Structural Genomics Consortium Target CgR113A | | Descriptor: | PYROPHOSPHATE, S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent methyltransferase | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Sahdev, S, Xiao, R, Foote, E.L, Ciccosanti, C, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-04-14 | | Release date: | 2009-04-28 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Northeast Structural Genomics Consortium Target CgR113A
To be published
|
|
3GL1
 
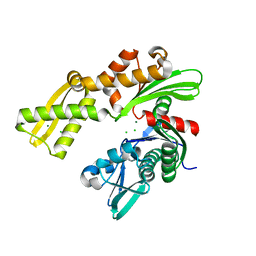 | | Crystal structure of ATPase domain of Ssb1 chaperone, a member of the HSP70 family, from Saccharomyces cerevisiae | | Descriptor: | CHLORIDE ION, GLYCEROL, Heat shock protein SSB1, ... | | Authors: | Osipiuk, J, Li, H, Bargassa, M, Sahi, C, Craig, E.A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-11 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of ATPase domain of Ssb1 chaperone, member of the HSP70 family from Saccharomyces cerevisiae.
To be Published
|
|
3GLU
 
 | | Crystal Structure of Human SIRT3 with AceCS2 peptide | | Descriptor: | Acetyl-coenzyme A synthetase 2-like, mitochondrial, NAD-dependent deacetylase sirtuin-3, ... | | Authors: | Jin, L, Wei, W, Jiang, Y, Peng, H, Cai, J, Mao, C, Dai, H, Bemis, J.E, Jirousek, M.R, Milne, J.C, Westphal, C.H, Perni, R.B. | | Deposit date: | 2009-03-12 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Human SIRT3 Displaying Substrate-induced Conformational Changes.
J.Biol.Chem., 284, 2009
|
|
3VOA
 
 | | Staphylococcus aureus FtsZ 12-316 GDP-form | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Yamane, J, Matsui, T, Mogi, N, Yao, M, Tanaka, I. | | Deposit date: | 2012-01-20 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural reorganization of the bacterial cell-division protein FtsZ from Staphylococcus aureus
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3VA8
 
 | | Crystal structure of enolase FG03645.1 (target EFI-502278) from Gibberella zeae PH-1 complexed with magnesium, formate and sulfate | | Descriptor: | FORMIC ACID, MAGNESIUM ION, PROBABLE DEHYDRATASE, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-12-29 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of dehydratase FG03645.1 from Gibberella zeae PH-1
To be Published
|
|
3VOT
 
 | | Crystal structure of L-amino acid ligase from Bacillus licheniformis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Suzuki, M, Takahashi, Y, Noguchi, A, Arai, T, Yagasaki, M, Kino, K, Saito, J. | | Deposit date: | 2012-02-08 | | Release date: | 2012-11-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of L-amino-acid ligase from Bacillus licheniformis
Acta Crystallogr.,Sect.D, 68, 2012
|
|
