7TYW
 
 | | Human Amylin1 Receptor in complex with Gs and salmon calcitonin peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Calcitonin receptor, ... | | Authors: | Cao, J, Belousoff, M.J, Johnson, R.M, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2022-02-14 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A structural basis for amylin receptor phenotype.
Science, 375, 2022
|
|
7TYO
 
 | | Calcitonin receptor in complex with Gs and human calcitonin peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Calcitonin, ... | | Authors: | Cao, J, Belousoff, M.J, Johnson, R.M, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2022-02-14 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A structural basis for amylin receptor phenotype.
Science, 375, 2022
|
|
7TYL
 
 | | Calcitonin Receptor in complex with Gs and rat amylin peptide, bypass motif | | Descriptor: | Calcitonin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cao, J, Belousoff, M.J, Johnson, R.M, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2022-02-13 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A structural basis for amylin receptor phenotype.
Science, 375, 2022
|
|
2NNC
 
 | | Structure of the sulfur carrier protein SoxY from Chlorobium limicola f thiosulfatophilum | | Descriptor: | CHLORIDE ION, NITROGEN MOLECULE, PHOSPHATE ION, ... | | Authors: | Stout, J, Van Driessche, G, Savvides, S.N, Van Beeumen, J. | | Deposit date: | 2006-10-24 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | X-ray crystallographic analysis of the sulfur carrier protein SoxY from Chlorobium limicola f. thiosulfatophilum reveals a tetrameric structure.
Protein Sci., 16, 2007
|
|
6OST
 
 | | RF2 pre-accommodated state bound Release complex 70S at 24ms | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-02 | | Release date: | 2019-06-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
4C69
 
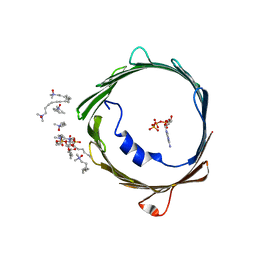 | | ATP binding to murine voltage-dependent anion channel 1 (mVDAC1). | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-5'-TRIPHOSPHATE, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Paz, A, Colletier, J.P, Abramson, J. | | Deposit date: | 2013-09-17 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.277 Å) | | Cite: | Structure-Guided Simulations Illuminate the Mechanism of ATP Transport Through Vdac1.
Nat.Struct.Mol.Biol., 21, 2014
|
|
7TYY
 
 | | Human Amylin2 Receptor in complex with Gs and salmon calcitonin peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Calcitonin receptor, ... | | Authors: | Cao, J, Belousoff, M.J, Johnson, R.M, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2022-02-14 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A structural basis for amylin receptor phenotype.
Science, 375, 2022
|
|
6OBF
 
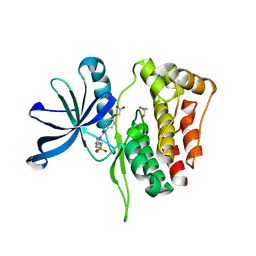 | | JAK2 JH2 in complex with JAK179 | | Descriptor: | GLYCEROL, Tyrosine-protein kinase JAK2, [4-({5-amino-3-[(4-sulfamoylphenyl)amino]-1H-1,2,4-triazole-1-carbonyl}amino)phenoxy]acetic acid | | Authors: | Krimmer, S.G, Liosi, M.E, Schlessinger, J, Jorgensen, W.L. | | Deposit date: | 2019-03-20 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Selective Janus Kinase 2 (JAK2) Pseudokinase Ligands with a Diaminotriazole Core.
J.Med.Chem., 63, 2020
|
|
5TF9
 
 | |
7TYN
 
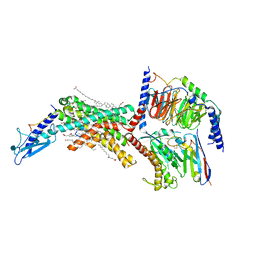 | | Calcitonin Receptor in complex with Gs and salmon calcitonin peptide | | Descriptor: | (2S)-2-{[(1R)-1-hydroxyhexadecyl]oxy}-3-{[(1R)-1-hydroxyoctadecyl]oxy}propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Cao, J, Belousoff, M.J, Johnson, R.M, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2022-02-13 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A structural basis for amylin receptor phenotype.
Science, 375, 2022
|
|
7TQB
 
 | |
7TQA
 
 | | Crystal Structure of monoclonal S9.6 Fab | | Descriptor: | Fab S9.6 heavy chain, Fab S9.6 light chain, GLYCEROL, ... | | Authors: | Bou-Nader, C, Zhang, J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-03-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.328 Å) | | Cite: | Structural basis of R-loop recognition by the S9.6 monoclonal antibody.
Nat Commun, 13, 2022
|
|
6OG0
 
 | | Structure of Aedes aegypti OBP22 | | Descriptor: | AAEL005772-PA, CADMIUM ION, CHLORIDE ION | | Authors: | Jones, D.N, Wang, J. | | Deposit date: | 2019-04-01 | | Release date: | 2019-04-17 | | Last modified: | 2020-05-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Aedes aegypti Odorant Binding Protein 22 selectively binds fatty acids through a conformational change in its C-terminal tail.
Sci Rep, 10, 2020
|
|
2W49
 
 | | ISOMETRICALLY CONTRACTING INSECT ASYNCHRONOUS FLIGHT MUSCLE | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, CALCIUM ION, ... | | Authors: | Wu, S, Liu, J, Reedy, M.C, Tregear, R.T, Winkler, H, Franzini-Armstrong, C, Sasaki, H, Lucaveche, C, Goldman, Y.E, Reedy, M.K, Taylor, K.A. | | Deposit date: | 2008-11-24 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | Electron Tomography of Cryofixed, Isometrically Contracting Insect Flight Muscle Reveals Novel Actin-Myosin Interactions
Plos One, 5, 2010
|
|
8APQ
 
 | | CaMct - Mesaconyl-CoA C1:C4 CoA Transferase of Chloroflexus aurantiacus | | Descriptor: | (2E)-2-METHYLBUT-2-ENEDIOIC ACID, 2-methylfumaryl-CoA isomerase, COENZYME A, ... | | Authors: | Pfister, P, Zarzycki, J, Erb, T.J. | | Deposit date: | 2022-08-10 | | Release date: | 2023-01-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural Basis for a Cork-Up Mechanism of the Intra-Molecular Mesaconyl-CoA Transferase.
Biochemistry, 62, 2023
|
|
4N1G
 
 | | Crystal Structure of Ca(2+)- discharged F88Y obelin mutant from Obelia longissima at 1.50 Angstrom resolution | | Descriptor: | CALCIUM ION, N-[3-BENZYL-5-(4-HYDROXYPHENYL)PYRAZIN-2-YL]-2-(4-HYDROXYPHENYL)ACETAMIDE, Obelin | | Authors: | Natashin, P.V, Markova, S.V, Lee, J, Vysotski, E.S, Liu, Z.J. | | Deposit date: | 2013-10-04 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of the F88Y obelin mutant before and after bioluminescence provide molecular insight into spectral tuning among hydromedusan photoproteins
Febs J., 281, 2014
|
|
7TYX
 
 | | Human Amylin2 Receptor in complex with Gs and rat amylin peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Calcitonin receptor, ... | | Authors: | Cao, J, Belousoff, M.J, Johnson, R.M, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2022-02-14 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | A structural basis for amylin receptor phenotype.
Science, 375, 2022
|
|
3EVX
 
 | | Crystal structure of the human E2-like ubiquitin-fold modifier conjugating enzyme 1 (Ufc1). Northeast Structural Genomics Consortium target HR41 | | Descriptor: | THIOCYANATE ION, Ufm1-conjugating enzyme 1 | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Ho, C.K, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-13 | | Release date: | 2008-10-21 | | Last modified: | 2023-01-04 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | NMR and X-RAY structures of human E2-like ubiquitin-fold modifier conjugating enzyme 1 (UFC1) reveal structural and functional conservation in the metazoan UFM1-UBA5-UFC1 ubiquination pathway.
J.STRUCT.FUNCT.GENOM., 10, 2009
|
|
8ATD
 
 | | Wild type hexamer oxalyl-CoA synthetase (OCS) | | Descriptor: | Oxalate--CoA ligase | | Authors: | Lill, P, Burgi, J, Raunser, S, Wilmanns, M, Gatsogiannis, C. | | Deposit date: | 2022-08-23 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Asymmetric horseshoe-like assembly of peroxisomal yeast oxalyl-CoA synthetase.
Biol.Chem., 404, 2023
|
|
5Y5E
 
 | | Crystal structure of phospholipase A2 with inhibitor | | Descriptor: | 2-[2-methyl-3-oxamoyl-1-[[2-(trifluoromethyl)phenyl]methyl]indol-4-yl]oxyethanoic acid, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-08-08 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2group IIE
Sci Rep, 7, 2017
|
|
7TYI
 
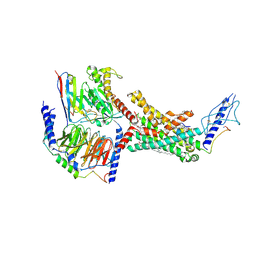 | | Calcitonin Receptor in complex with Gs and rat amylin peptide, CT-like state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Calcitonin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cao, J, Belousoff, M.J, Johnson, R.M, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2022-02-13 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A structural basis for amylin receptor phenotype.
Science, 375, 2022
|
|
4C5W
 
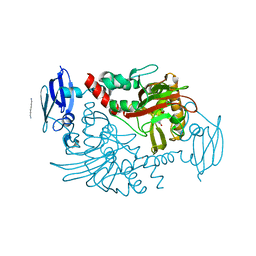 | | Three dimensional structure of human gamma-butyrobetaine hydroxylase in complex with 4-(Ethyldimethylammonio)butanoate | | Descriptor: | GAMMA-BUTYROBETAINE DIOXYGENASE, HEXANE-1,6-DIAMINE, N-OXALYLGLYCINE, ... | | Authors: | Tars, K, Leitans, J, Kazaks, A. | | Deposit date: | 2013-09-16 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Targeting Carnitine Biosynthesis: Discovery of New Inhibitors Against Gamma-Butyrobetaine Hydroxylase.
J.Med.Chem., 57, 2014
|
|
8AFF
 
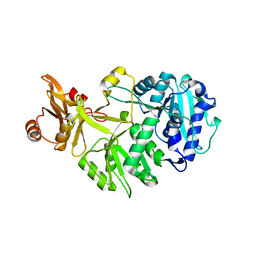 | |
2NB7
 
 | |
3WUB
 
 | | The wild type crystal structure of b-1,4-Xylanase (XynAS9) from Streptomyces sp. 9 | | Descriptor: | Endo-1,4-beta-xylanase A, ZINC ION | | Authors: | Chen, C.C, Han, X, Lv, P, Ko, T.P, Peng, W, Huang, C.H, Zheng, Y, Gao, J, Yang, Y.Y, Guo, R.T. | | Deposit date: | 2014-04-23 | | Release date: | 2014-10-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural perspectives of an engineered beta-1,4-xylanase with enhanced thermostability.
J.Biotechnol., 189C, 2014
|
|
