3D8B
 
 | | Crystal structure of human fidgetin-like protein 1 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Fidgetin-like protein 1 | | Authors: | Karlberg, T, Wisniewska, M, Andersson, J, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Olesen, K, Persson, C, Sagemark, J, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Welin, M, Wikstrom, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-23 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human fidgetin-like protein 1.
To be Published
|
|
6Z71
 
 | | Structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | Descriptor: | Aq128 | | Authors: | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | Deposit date: | 2020-05-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of the Aquifex aeolicus MATE family multidrug resistance transporter and sequence comparisons suggest the existence of a new subfamily.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6Z70
 
 | | Structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Aq128 | | Authors: | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | Deposit date: | 2020-05-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the Aquifex aeolicus MATE family multidrug resistance transporter and sequence comparisons suggest the existence of a new subfamily.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3DG4
 
 | |
4YYA
 
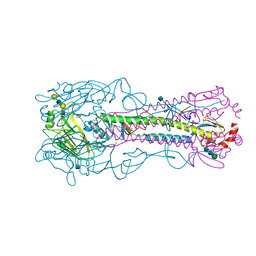 | | The structure of hemagglutinin from a H6N1 influenza virus (A/Taiwan/2/2013) in complex with avian receptor analog 3'SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, HA2, ... | | Authors: | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-03-23 | | Release date: | 2016-04-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013)
To Be Published
|
|
6U2P
 
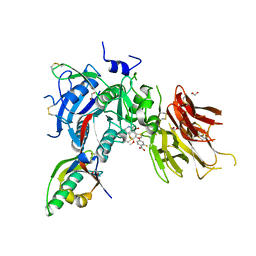 | | PCSK9 in complex with compound 5 | | Descriptor: | 1,2-ETHANEDIOL, 2-fluoro-4-{[(1R)-6-methoxy-1-methyl-1-{2-oxo-2-[(1,3-thiazol-2-yl)amino]ethyl}-1,2,3,4-tetrahydroisoquinolin-7-yl]oxy}benzoic acid, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Lu, J, Soisson, S. | | Deposit date: | 2019-08-20 | | Release date: | 2019-11-06 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | From Screening to Targeted Degradation: Strategies for the Discovery and Optimization of Small Molecule Ligands for PCSK9.
Cell Chem Biol, 27, 2020
|
|
3D8F
 
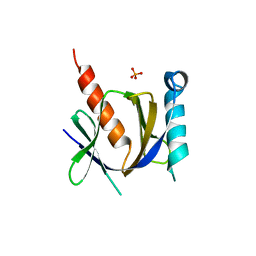 | | Crystal structure of the human Fe65-PTB1 domain with bound phosphate (trigonal crystal form) | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 1, PHOSPHATE ION | | Authors: | Radzimanowski, J, Ravaud, S, Sinning, I, Wild, K. | | Deposit date: | 2008-05-23 | | Release date: | 2008-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the human Fe65-PTB1 domain.
J.Biol.Chem., 283, 2008
|
|
3DJB
 
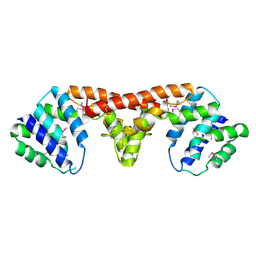 | | Crystal structure of a HD-superfamily hydrolase (BT9727_1981) from Bacillus thuringiensis, Northeast Structural Genomics Consortium Target BuR114 | | Descriptor: | Hydrolase, HD family, MAGNESIUM ION | | Authors: | Forouhar, F, Su, M, Seetharaman, J, Vorobiev, S.M, Janjua, H, Fang, Y, Xiao, R, Cunningham, K, Maglaqui, M, Owen, L.A, Wang, D, Baran, M.C, Liu, J, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-23 | | Release date: | 2008-08-19 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a HD-superfamily hydrolase (BT9727_1981) from Bacillus thuringiensis, Northeast Structural Genomics Consortium Target BuR114
To be Published
|
|
3D8D
 
 | | Crystal structure of the human Fe65-PTB1 domain | | Descriptor: | 1,2-ETHANEDIOL, Amyloid beta A4 precursor protein-binding family B member 1, MERCURY (II) ION | | Authors: | Radzimanowski, J, Ravaud, S, Sinning, I, Wild, K. | | Deposit date: | 2008-05-23 | | Release date: | 2008-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the human Fe65-PTB1 domain.
J.Biol.Chem., 283, 2008
|
|
7AAZ
 
 | | Crystal structure of MerTK in complex with a type 1.5 aminopyridine inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-azanyl-~{N}-[(1~{S},2~{S})-2-[[4-[4-[(4-methylpiperazin-1-yl)methyl]phenyl]phenyl]methoxy]cyclopentyl]-5-(1-methylpyrazol-4-yl)pyridine-3-carboxamide, CHLORIDE ION, ... | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Overman, R.C, Rawlins, P.B, Truman, C, Underwood, E, Warwicker, J, Winter-Holt, J. | | Deposit date: | 2020-09-05 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | A-loop interactions in Mer tyrosine kinase give rise to inhibitors with two-step mechanism and long residence time of binding.
Biochem.J., 477, 2020
|
|
3DG0
 
 | |
6ZPH
 
 | | Kinesin binding protein complexed with Kif15 motor domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KIF-binding protein, Kinesin-like protein KIF15, ... | | Authors: | Atherton, J, Hummel, J.J.A, Olieric, N, Locke, J, Pena, A, Rosenfeld, S.S, Steinmetz, M.O, Hoogenraad, C.C, Moores, C.A. | | Deposit date: | 2020-07-08 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | The mechanism of kinesin inhibition by kinesin-binding protein.
Elife, 9, 2020
|
|
3DH1
 
 | | Crystal structure of human tRNA-specific adenosine-34 deaminase subunit ADAT2 | | Descriptor: | ZINC ION, tRNA-specific adenosine deaminase 2 | | Authors: | Welin, M, Tresaugues, L, Andersson, J, Arrowsmith, C.H, Berglund, H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nyman, T, Olesen, K, Persson, C, Sagemark, J, Schueler, H, Thorsell, A.G, van der Berg, S, Wisniewska, M, Wikstrom, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-16 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human tRNA-specific adenosine-34 deaminase subunit ADAT2.
To be Published
|
|
6ZPI
 
 | | Microtubule complexed with Kif15 motor domain. Symmetrised asymmetric unit | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF15, ... | | Authors: | Atherton, J, Hummel, J.J.A, Olieric, N, Locke, J, Pena, A, Rosenfeld, S.S, Steinmetz, M.O, Hoogenraad, C.C, Moores, C.A. | | Deposit date: | 2020-07-08 | | Release date: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The mechanism of kinesin inhibition by kinesin-binding protein.
Elife, 9, 2020
|
|
6ZWC
 
 | | Z-SBTub2 photoswitch bound to tubulin-DARPin D1 complex | | Descriptor: | 2-[2-(3,4,5-trimethoxyphenyl)ethyl]-1,3-benzothiazole, Designed Ankyrin Repeat Protein (DARPIN) D1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wranik, M, Weinert, T, Olieric, N, Gao, L, Kraus, Y.C.M, Bingham, R, Ntouliou, E, Ahlfeld, J, Thorn-Seshold, O, Steinmetz, M.O, Standfuss, J. | | Deposit date: | 2020-07-28 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | A Robust, GFP-Orthogonal Photoswitchable Inhibitor Scaffold Extends Optical Control over the Microtubule Cytoskeleton.
Cell Chem Biol, 28, 2021
|
|
6R0J
 
 | |
8B24
 
 | | Time-resolved structure of K+-dependent Na+-PPase from Thermotoga maritima 3600-seconds post reaction initiation with Na+ | | Descriptor: | DIPHOSPHATE, K(+)-stimulated pyrophosphate-energized sodium pump, MAGNESIUM ION, ... | | Authors: | Strauss, J, Vidilaseris, K, Goldman, A. | | Deposit date: | 2022-09-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (4.53 Å) | | Cite: | Functional and structural asymmetry suggest a unifying principle for catalysis in membrane-bound pyrophosphatases.
Embo Rep., 25, 2024
|
|
8B22
 
 | |
8B23
 
 | |
8B21
 
 | | Time-resolved structure of K+-dependent Na+-PPase from Thermotoga maritima 0-60-seconds post reaction initiation with Na+ | | Descriptor: | DI(HYDROXYETHYL)ETHER, DODECYL-BETA-D-MALTOSIDE, K(+)-stimulated pyrophosphate-energized sodium pump, ... | | Authors: | Strauss, J, Vidilaseris, K, Goldman, A. | | Deposit date: | 2022-09-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Functional and structural asymmetry suggest a unifying principle for catalysis in membrane-bound pyrophosphatases.
Embo Rep., 25, 2024
|
|
3DX9
 
 | | Crystal Structure of the DM1 TCR at 2.75A | | Descriptor: | DM1 T cell receptor alpha chain, DM1 T cell receptor beta chain | | Authors: | Archbold, J.K, Macdonald, W.A, Gras, S, Rossjohn, J. | | Deposit date: | 2008-07-24 | | Release date: | 2009-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Natural micropolymorphism in human leukocyte antigens provides a basis for genetic control of antigen recognition.
J.Exp.Med., 206, 2009
|
|
3DME
 
 | | Crystal structure of conserved exported protein from Bordetella pertussis. NorthEast Structural Genomics target BeR141 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L(+)-TARTARIC ACID, conserved exported protein | | Authors: | Seetharaman, J, Abashidze, M, Wang, D, Janjua, H, Foote, E.L, Xiao, R, Liu, J, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-30 | | Release date: | 2008-08-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of conserved exported protein from Bordetella pertussis. NorthEast Structural Genomics target BeR141
To be Published
|
|
6SKK
 
 | | Structure of the native full-length HIV-1 capsid protein in helical assembly (-13,8) | | Descriptor: | capsid protein | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2019-08-15 | | Release date: | 2020-08-26 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7AAX
 
 | | Crystal structure of MerTK kinase domain in complex with LDC1267 | | Descriptor: | CHLORIDE ION, Tyrosine-protein kinase Mer, ~{N}-[4-(6,7-dimethoxyquinolin-4-yl)oxy-3-fluoranyl-phenyl]-4-ethoxy-1-(4-fluoranyl-2-methyl-phenyl)pyrazole-3-carboxamide | | Authors: | Schimpl, M, Pflug, A, McCoull, W, Nissink, J.W.M, Overman, R.C, Rawlins, P.B, Truman, C, Underwood, E, Warwicker, J, Winter-Holt, J. | | Deposit date: | 2020-09-05 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | A-loop interactions in Mer tyrosine kinase give rise to inhibitors with two-step mechanism and long residence time of binding.
Biochem.J., 477, 2020
|
|
7AB0
 
 | | Apo crystal structure of the MerTK kinase domain | | Descriptor: | CHLORIDE ION, Tyrosine-protein kinase Mer | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Overman, R.C, Rawlins, P.B, Truman, C, Underwood, E, Warwicker, J, Winter-Holt, J. | | Deposit date: | 2020-09-05 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A-loop interactions in Mer tyrosine kinase give rise to inhibitors with two-step mechanism and long residence time of binding.
Biochem.J., 477, 2020
|
|
