5ZVQ
 
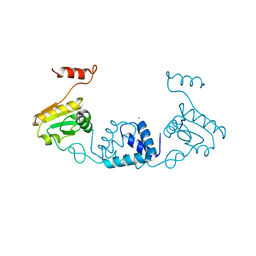 | |
5PCZ
 
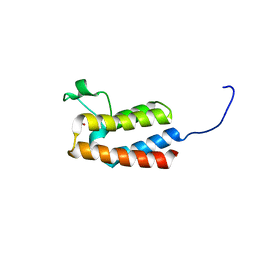 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 55) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
6E0I
 
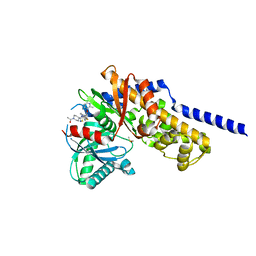 | | Crystal structure of Glucokinase in complex with compound 72 | | Descriptor: | 1-{4-[5-({3-[(2-methylpyridin-3-yl)oxy]-5-[(pyridin-2-yl)sulfanyl]pyridin-2-yl}amino)-1,2,4-thiadiazol-3-yl]piperidin-1 -yl}ethan-1-one, DIMETHYL SULFOXIDE, Glucokinase, ... | | Authors: | Hinklin, R.J, Baer, B.R, Boyd, S.A, Chicarelli, M.D, Condroski, K.R, DeWolf, W.E, Fischer, J, Frank, M, Hingorani, G.P, Lee, P.A, Neitzel, N.A, Pratt, S.A, Singh, A, Sullivan, F.X, Turner, T, Voegtli, W.C, Wallace, E.M, Williams, L, Aicher, T.D. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and preclinical development of AR453588 as an anti-diabetic glucokinase activator.
Bioorg.Med.Chem., 28, 2020
|
|
6A5M
 
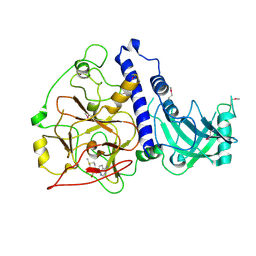 | | Crystal structure of Arabidopsis thaliana SUVH6 in complex with SAM, form 2 | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH6, S-ADENOSYLMETHIONINE, ... | | Authors: | Li, X, Du, J. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2FIV
 
 | | Crystal structure of feline immunodeficiency virus protease complexed with a substrate | | Descriptor: | ACE-ALN-VAL-STA-GLU-ALN-NH2, FELINE IMMUNODEFICIENCY VIRUS PROTEASE, SULFATE ION | | Authors: | Schalk-Hihi, C, Lubkowski, J, Zdanov, A, Wlodawer, A, Gustchina, A. | | Deposit date: | 1997-07-21 | | Release date: | 1997-11-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the inactive D30N mutant of feline immunodeficiency virus protease complexed with a substrate and an inhibitor.
Biochemistry, 36, 1997
|
|
6DYK
 
 | |
5PDF
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 71) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
6A7C
 
 | | Human dihydrofolate reductase complexed with NADPH and BT1 | | Descriptor: | 5-(3-{3-[(2,4-diamino-6-ethylpyrimidin-5-yl)oxy]propoxy}phenyl)-6-ethylpyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Vanichtanankul, J, Tarnchompoo, B, Chitnumsub, P, Kamchonwongpaisan, S, Yuthavong, Y. | | Deposit date: | 2018-07-02 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Hybrid Inhibitors of Malarial Dihydrofolate Reductase with Dual Binding Modes That Can Forestall Resistance.
ACS Med Chem Lett, 9, 2018
|
|
6DY4
 
 | |
3E5V
 
 | | Crystal Structure Analysis of eqFP611 Double Mutant T122R, N143S | | Descriptor: | Red fluorescent protein eqFP611 | | Authors: | Nar, H, Nienhaus, K, Nienhaus, U, Wiedenmann, J. | | Deposit date: | 2008-08-14 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Trans-cis isomerization is responsible for the red-shifted fluorescence in variants of the red fluorescent protein eqFP611.
J.Am.Chem.Soc., 130, 2008
|
|
6DY8
 
 | |
3E61
 
 | | Crystal structure of a putative transcriptional repressor of ribose operon from Staphylococcus saprophyticus subsp. saprophyticus | | Descriptor: | GLYCEROL, Putative transcriptional repressor of ribose operon | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-14 | | Release date: | 2008-08-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative transcriptional repressor of ribose operon from Staphylococcus saprophyticus subsp. saprophyticus
To be Published
|
|
6DYI
 
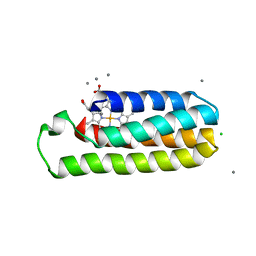 | | Co(II)-bound structure of the engineered cyt cb562 variant, H3 | | Descriptor: | CALCIUM ION, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Tezcan, F.A, Rittle, J. | | Deposit date: | 2018-07-01 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.964 Å) | | Cite: | An efficient, step-economical strategy for the design of functional metalloproteins.
Nat.Chem., 11, 2019
|
|
6A8L
 
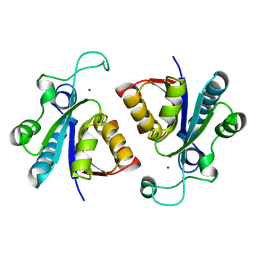 | | Crystal structure of nicotinamidase/ pyrazinamidase PncA from Bacillus subtilis | | Descriptor: | Isochorismatase, ZINC ION | | Authors: | Shang, F, Chen, J, Wang, L, Xu, Y. | | Deposit date: | 2018-07-09 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the nicotinamidase/pyrazinamidase PncA from Bacillus subtilis.
Biochem. Biophys. Res. Commun., 503, 2018
|
|
3E6Q
 
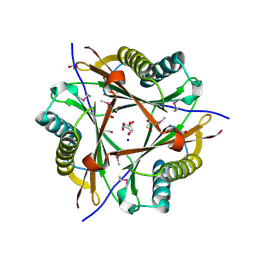 | | Putative 5-carboxymethyl-2-hydroxymuconate isomerase from Pseudomonas aeruginosa. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FORMIC ACID, ... | | Authors: | Osipiuk, J, Xu, X, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-08-15 | | Release date: | 2008-08-26 | | Last modified: | 2020-05-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | X-ray crystal structure of putative 5-carboxymethyl-2-hydroxymuconate isomerase from Pseudomonas aeruginosa.
To be Published
|
|
3EBK
 
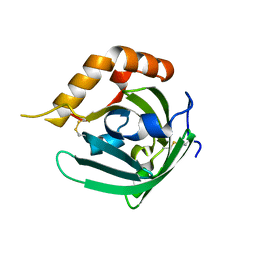 | | Crystal structure of major allergens, Bla g 4 from cockroaches | | Descriptor: | Allergen Bla g 4 | | Authors: | Tan, Y.W, Chan, S.L, Chew, F.T, Sivaraman, J, Mok, Y.K. | | Deposit date: | 2008-08-28 | | Release date: | 2008-12-02 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of two major allergens, Bla g 4 and Per a 4, from cockroaches and their IgE binding epitopes.
J.Biol.Chem., 284, 2008
|
|
6DYB
 
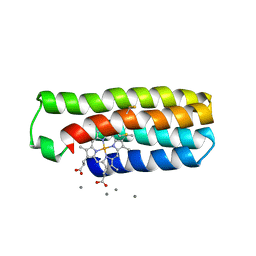 | |
6DYJ
 
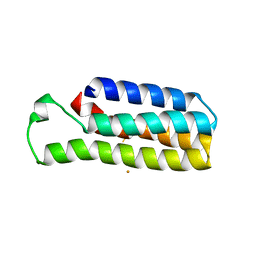 | |
6DYO
 
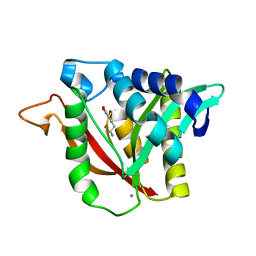 | | C-terminal condensation domain of Ebony in complex with L-Dopamine | | Descriptor: | CALCIUM ION, Ebony, L-DOPAMINE | | Authors: | Izore, T, Tailhades, J, Hansen, M.H, Kaczmarski, J.A, Jackson, C.J, Cryle, M.J. | | Deposit date: | 2018-07-02 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Drosophila melanogasternonribosomal peptide synthetase Ebony encodes an atypical condensation domain.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
3EN2
 
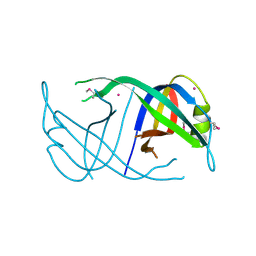 | | Three-dimensional structure of the protein priB from Ralstonia solanacearum at the resolution 2.3A. Northeast Structural Genomics Consortium target RsR213C. | | Descriptor: | POTASSIUM ION, Probable primosomal replication protein n | | Authors: | Kuzin, A.P, Neely, H, Wang, H, Sahdev, S, Foote, E.L, Xiao, R, Liu, J, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of the protein priB from Ralstonia solanacearum at the resolution 2.3A. Northeast Structural Genomics Consortium target RsR213C.
To be Published
|
|
5OVU
 
 | | Cupriavidus metallidurans BPH | | Descriptor: | BETA-PROTEOBACTERIA PROTEASOME HOMOLOGUE, MALONATE ION | | Authors: | Fuchs, A.C.D, Albrecht, R, Martin, J, Hartmann, M.D. | | Deposit date: | 2017-08-29 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of the bacterial proteasome homolog BPH reveals a tetradecameric double-ring complex with unique inner cavity properties.
J. Biol. Chem., 293, 2018
|
|
6E1R
 
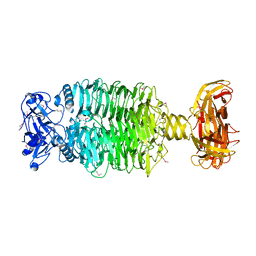 | | Crystal structure of the Acinetobacter phage vB_ApiP_P1 tailspike protein | | Descriptor: | CHLORIDE ION, SODIUM ION, Tailspike protein | | Authors: | Plattner, M, Shneider, M.M, Oliveira, H, Azeredo, J, Leiman, P.G. | | Deposit date: | 2018-07-10 | | Release date: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Crystal structure of the Acinetobacter phage vB_ApiP_P1 tailspike protein
To Be Published
|
|
6DYD
 
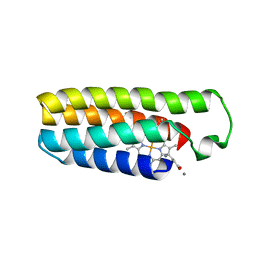 | | Cu(II)-bound structure of the engineered cyt cb562 variant, CH3 | | Descriptor: | CALCIUM ION, COPPER (II) ION, HEME C, ... | | Authors: | Tezcan, F.A, Rittle, J. | | Deposit date: | 2018-07-01 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.724 Å) | | Cite: | An efficient, step-economical strategy for the design of functional metalloproteins.
Nat.Chem., 11, 2019
|
|
6DYM
 
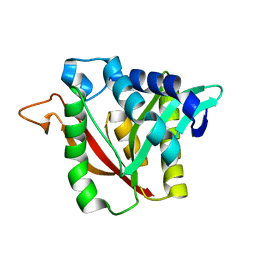 | | C-terminal condensation domain of Ebony | | Descriptor: | CALCIUM ION, Ebony | | Authors: | Izore, T, Tailhades, J, Hansen, M.H, Kaczmarski, J.A, Jackson, C.J, Cryle, M.J. | | Deposit date: | 2018-07-02 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Drosophila melanogasternonribosomal peptide synthetase Ebony encodes an atypical condensation domain.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
3E2W
 
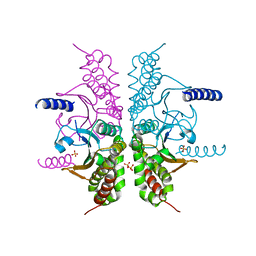 | |
