6Y2M
 
 | | Streptavidin mutant S112R with a biotC4-1 cofactor - an artificial iron hydroxylase | | Descriptor: | Streptavidin, biotC4-1 cofactor | | Authors: | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | Deposit date: | 2020-02-17 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
6ZN2
 
 | | Partial structure of tyrosine hydroxylase in complex with dopamine showing the catalytic domain and an alpha-helix from the regulatory domain involved in dopamine binding. | | Descriptor: | FE (III) ION, L-DOPAMINE, SER-LEU-ILE-GLU-ASP-ALA-ARG-LYS-GLU-ARG-GLU-ALA-ALA-VAL-ALA-ALA-ALA-ALA, ... | | Authors: | Bueno-Carrasco, M.T, Cuellar, J, Santiago, C, Valpuesta, J.M, Martinez, A, Flydal, M.I. | | Deposit date: | 2020-07-06 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation.
Nat Commun, 13, 2022
|
|
6Y2T
 
 | | Streptavidin wildtype with a biotC4-1 cofactor - an artificial iron hydroxylase | | Descriptor: | GLYCEROL, Streptavidin, biotC4-1 cofactor | | Authors: | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | Deposit date: | 2020-02-17 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
3L3K
 
 | | Crystal structure of HLA-B*4402 in complex with the R5A/F7A double mutant of a self-peptide derived from DPA*0201 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Theodossis, A, Ely, L.K, Rossjohn, J. | | Deposit date: | 2009-12-17 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Constraints within major histocompatibility complex class I restricted peptides: presentation and consequences for T-cell recognition
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1KLM
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH BHAP U-90152 | | Descriptor: | (1-(5-METHANSULPHONAMIDO-1H-INDOL-2-YL-CARBONYL)4-[METHYLAMINO)PYRIDINYL]PIPERAZINE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R.M, Stammers, D.K, Stuart, D.I. | | Deposit date: | 1997-03-17 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Unique features in the structure of the complex between HIV-1 reverse transcriptase and the bis(heteroaryl)piperazine (BHAP) U-90152 explain resistance mutations for this nonnucleoside inhibitor.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
6ZZU
 
 | | Partial structure of the substrate-free tyrosine hydroxylase (apo-TH). | | Descriptor: | FE (III) ION, Tyrosine 3-monooxygenase | | Authors: | Bueno-Carrasco, M.T, Cuellar, J, Santiago, C, Valpuesta, J.M, Martinez, A, Flydal, M.I. | | Deposit date: | 2020-08-05 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation.
Nat Commun, 13, 2022
|
|
7A2G
 
 | | Full-length structure of the substrate-free tyrosine hydroxylase (apo-TH). | | Descriptor: | FE (III) ION, Tyrosine 3-monooxygenase | | Authors: | Bueno-Carrasco, M.T, Cuellar, J, Santiago, C, Flydal, M.I, Martinez, A, Valpuesta, J.M. | | Deposit date: | 2020-08-17 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation.
Nat Commun, 13, 2022
|
|
5V9I
 
 | | Crystal structure of catalytic domain of G9a with MS0105 | | Descriptor: | GLYCEROL, Histone-lysine N-methyltransferase EHMT2, N~2~-cyclohexyl-N~4~-(1-ethylpiperidin-4-yl)-6,7-dimethoxy-N~2~-methylquinazoline-2,4-diamine, ... | | Authors: | Dong, A, Zeng, H, Liu, J, Xiong, Y, Babault, N, Jin, J, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Wu, H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-03-23 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of catalytic domain of G9a with MS0105
to be published
|
|
6Y3Q
 
 | | Streptavidin mutant S112R_K121E with a biotC5-1 cofactor - an artificial iron hydroxylase | | Descriptor: | SULFATE ION, Streptavidin, biotC5-1 cofactor | | Authors: | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | Deposit date: | 2020-02-18 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
3LPI
 
 | | Structure of BACE Bound to SCH745132 | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N'-{(1S,2S)-1-(3,5-difluorobenzyl)-2-hydroxy-2-[(2R)-4-(phenylsulfonyl)piperazin-2-yl]ethyl}-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2010-02-05 | | Release date: | 2010-04-14 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4DWI
 
 | | Crystal structure of fragment DNA polymerase I from Bacillus stearothermophilus with self complementary DNA, Se-dGTP and Calcium | | Descriptor: | 9-METHYLGUANINE, CALCIUM ION, DNA (5'-D(*AP*CP*TP*GP*GP*AP*TP*CP*CP*A)-3'), ... | | Authors: | Gan, J.H, Abdur, R, Liu, H.H, Sheng, J, Caton-Willians, J, Soares, A.S, Huang, Z. | | Deposit date: | 2012-02-24 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical and structural insights into the fidelity of bacillus stearothermophilus DNA polymerase
To be Published
|
|
7ADZ
 
 | | Cryo-EM structure of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis, the cap portion in extended state. | | Descriptor: | Phage tail protein, Putative phage tail sheath protein FI, cap adaptor protein (Algo2), ... | | Authors: | Xu, J, Ericson, C, Feldmueller, M, Lien, Y.W, Pilhofer, M. | | Deposit date: | 2020-09-17 | | Release date: | 2022-02-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Identification and structure of an extracellular contractile injection system from the marine bacterium Algoriphagus machipongonensis.
Nat Microbiol, 7, 2022
|
|
7AEB
 
 | | Cryo-EM structure of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis, the baseplate complex in extended state applied 6-fold symmetry. | | Descriptor: | Baseplate_J domain-containing protein, LysM domain-containing protein, Phage tail protein, ... | | Authors: | Xu, J, Ericson, C, Feldmueller, M, Lien, Y.W, Pilhofer, M. | | Deposit date: | 2020-09-17 | | Release date: | 2022-02-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Identification and structure of an extracellular contractile injection system from the marine bacterium Algoriphagus machipongonensis.
Nat Microbiol, 7, 2022
|
|
4DYL
 
 | | F-BAR domain of human FES tyrosine kinase | | Descriptor: | Tyrosine-protein kinase Fes/Fps | | Authors: | Ugochukwu, E, Salah, E, Elkins, J, Barr, A, Krojer, T, Filippakopoulos, P, Weigelt, J, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-02-29 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | F-BAR domain of human FES tyrosine kinase
TO BE PUBLISHED
|
|
7AE0
 
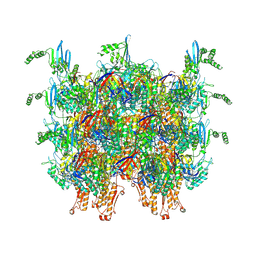 | | Cryo-EM structure of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis, the sheath-tube module in extended state. | | Descriptor: | Phage tail protein, Putative phage tail sheath protein FI | | Authors: | Xu, J, Ericson, C, Feldmueller, M, Lien, Y.W, Pilhofer, M. | | Deposit date: | 2020-09-17 | | Release date: | 2022-02-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Identification and structure of an extracellular contractile injection system from the marine bacterium Algoriphagus machipongonensis.
Nat Microbiol, 7, 2022
|
|
6Y9E
 
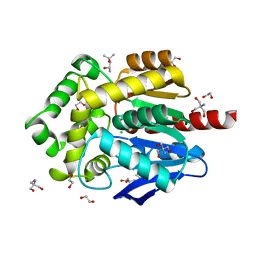 | | Crystal structure of putative ancestral haloalkane dehalogenase AncHLD2 (node 2) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Chaloupkova, R, Damborsky, J, Marek, M. | | Deposit date: | 2020-03-09 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of hyperstable ancestral haloalkane dehalogenases show restricted conformational dynamics.
Comput Struct Biotechnol J, 18, 2020
|
|
7AEF
 
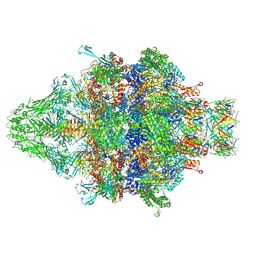 | | Cryo-EM structure of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis, the baseplate complex in extended state applied 3-fold symmetry. | | Descriptor: | Baseplate_J domain-containing protein, LysM domain-containing protein, Phage tail protein, ... | | Authors: | Xu, J, Ericson, C, Feldmueller, M, Lien, Y.W, Pilhofer, M. | | Deposit date: | 2020-09-17 | | Release date: | 2022-02-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Identification and structure of an extracellular contractile injection system from the marine bacterium Algoriphagus machipongonensis.
Nat Microbiol, 7, 2022
|
|
2AOJ
 
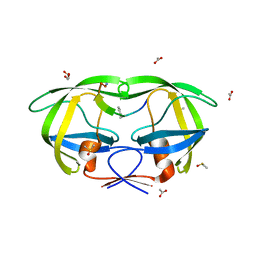 | | Crystal structure analysis of HIV-1 protease with a substrate analog P6-PR | | Descriptor: | ACETIC ACID, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
1QNY
 
 | | X-ray refinement of D2O soaked crystal of concanavalin A | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION | | Authors: | Habash, J, Raftery, J, Nuttall, R, Price, H.J, Lehmann, M.S, Wilkinson, C, Kalb(Gilboa), A.J, Helliwell, J.R. | | Deposit date: | 1999-10-26 | | Release date: | 2000-04-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct Determination of the Positions of Deuterium Atoms of Bound Water in Concanavalin a by Neutron Laue Crystallography
Acta Crystallogr.,Sect.D, 56, 2000
|
|
6ZSU
 
 | |
6ZSV
 
 | | Structure of crocagin biosynthetic protein CgnB | | Descriptor: | Uncharacterized protein, ZINC ION | | Authors: | Koehnke, J, Adam, S. | | Deposit date: | 2020-07-16 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Unusual peptide-binding proteins guide pyrroloindoline alkaloid formation in crocagin biosynthesis.
Nat.Chem., 15, 2023
|
|
7A0O
 
 | | NMR structure of flagelliform spidroin (FlagSp) N-terminal domain from Trichonephila clavipes at pH 5.5 | | Descriptor: | Flagelliform spidroin variant 1 | | Authors: | Sarr, M, Kitoka, K, Walsh-White, K.-A, Kaldmae, M, Landreh, M, Rising, A, Johansson, J, Jaudzems, K, Kronqvist, N. | | Deposit date: | 2020-08-10 | | Release date: | 2021-08-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The dimerization mechanism of the N-terminal domain of spider silk proteins is conserved despite extensive sequence divergence.
J.Biol.Chem., 298, 2022
|
|
6ZX6
 
 | |
5V9U
 
 | | Crystal Structure of small molecule ARS-1620 covalently bound to K-Ras G12C | | Descriptor: | (S)-1-{4-[6-chloro-8-fluoro-7-(2-fluoro-6-hydroxyphenyl)quinazolin-4-yl] piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | Authors: | Janes, M.R, Zhang, J, Li, L.-S, Hansen, R, Peters, U, Guo, X, Chen, Y, Babbar, A, Firdaus, S.J, Feng, J, Chen, J.H, Li, S, Brehmer, D, Darjania, L, Li, S, Long, Y.O, Thach, C, Liu, Y, Zarieh, A, Ely, T, Kucharski, J.M, Kessler, L.V, Wu, T, Wang, Y, Yao, Y, Deng, X, Zarrinkar, P, Dashyant, D, Lorenzi, M.V, Hu-Lowe, D, Patricelli, M.P, Ren, P, Liu, Y. | | Deposit date: | 2017-03-23 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor.
Cell, 172, 2018
|
|
6ZX7
 
 | |
