4ECI
 
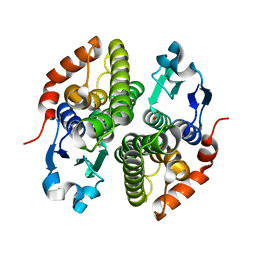 | | Crystal structure of glutathione s-transferase prk13972 (target efi-501853) from pseudomonas aeruginosa pacs2 complexed with acetate | | Descriptor: | ACETATE ION, glutathione S-transferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-26 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase Prk13972 from Pseudomonas Aeruginosa
To be Published
|
|
4E4F
 
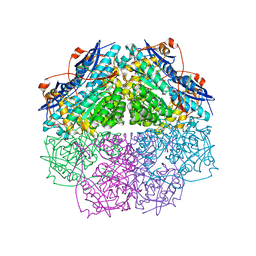 | | Crystal structure of enolase PC1_0802 (TARGET EFI-502240) from Pectobacterium carotovorum subsp. carotovorum PC1 | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-12 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of ENOLASE PC1_0802 from Pectobacterium carotovorum
To be Published
|
|
4E6U
 
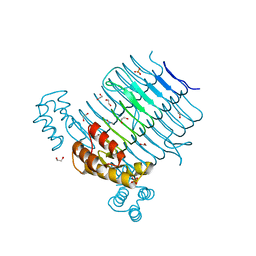 | | Structure of LpxA from Acinetobacter baumannii at 1.4A resolution (P63 form) | | Descriptor: | 1,2-ETHANEDIOL, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, SULFATE ION | | Authors: | Badger, J, Chie-Leon, B, Logan, C, Sridhar, V, Sankaran, B, Zwart, P.H, Nienaber, V. | | Deposit date: | 2012-03-16 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structure determination of LpxA from the lipopolysaccharide-synthesis pathway of Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4EHD
 
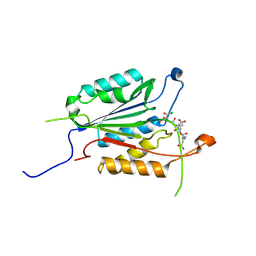 | | Allosteric Modulation of Caspase-3 through Mutagenesis | | Descriptor: | ACE-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE INHIBITOR, Caspase-3 | | Authors: | Walters, J, Schipper, J.L, Swartz, P, Mattos, C, Clark, A.C. | | Deposit date: | 2012-04-02 | | Release date: | 2012-06-06 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Allosteric modulation of caspase 3 through mutagenesis.
Biosci.Rep., 32, 2012
|
|
1ULZ
 
 | | Crystal structure of the biotin carboxylase subunit of pyruvate carboxylase | | Descriptor: | pyruvate carboxylase n-terminal domain | | Authors: | Kondo, S, Nakajima, Y, Sugio, S, Yong-Biao, J, Sueda, S, Kondo, H. | | Deposit date: | 2003-09-18 | | Release date: | 2004-03-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the biotin carboxylase subunit of pyruvate carboxylase from Aquifex aeolicus at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
4ELM
 
 | | Crystal structure of the mouse CD1d-lysosulfatide-Hy19.3 TCR complex | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl 3-O-sulfo-beta-D-galactopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Girardi, E, Maricic, I, Wang, J, Mac, T.T, Iyer, P, Kumar, V, Zajonc, D.M. | | Deposit date: | 2012-04-11 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Type II natural killer T cells use features of both innate-like and conventional T cells to recognize sulfatide self antigens.
Nat.Immunol., 13, 2012
|
|
4EHN
 
 | | Allosteric Modulation of Caspase-3 through Mutagenesis | | Descriptor: | ACE-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE INHIBITOR, Caspase-3 | | Authors: | Walters, J, Schipper, J.L, Swartz, P.D, Mattos, C, Clark, A.C. | | Deposit date: | 2012-04-02 | | Release date: | 2012-06-06 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Allosteric modulation of caspase 3 through mutagenesis.
Biosci.Rep., 32, 2012
|
|
1UTO
 
 | | Trypsin specificity as elucidated by LIE calculations, X-ray structures and association constant measurements | | Descriptor: | 2-PHENYLETHYLAMINE, CALCIUM ION, GLYCEROL, ... | | Authors: | Leiros, H.-K.S, Brandsdal, B.O, Andersen, O.A, Os, V, Leiros, I, Helland, R, Otlewski, J, Willassen, N.P, Smalas, A.O. | | Deposit date: | 2003-12-09 | | Release date: | 2004-01-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Trypsin Specificity as Elucidated by Lie Calculations, X-Ray Structures, and Association Constant Measurements
Protein Sci., 13, 2004
|
|
4EO1
 
 | | crystal structure of the TolA binding domain from the filamentous phage IKe | | Descriptor: | Attachment protein G3P, MAGNESIUM ION | | Authors: | Jakob, R.P, Geitner, A.J, Weininger, U, Balbach, J, Dobbek, H, Schmid, F.X. | | Deposit date: | 2012-04-13 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and energetic basis of infection by the filamentous bacteriophage IKe.
Mol.Microbiol., 84, 2012
|
|
4EW5
 
 | | C-terminal domain of inner membrane protein CigR from Salmonella enterica. | | Descriptor: | 1,2-ETHANEDIOL, CigR Protein | | Authors: | Osipiuk, J, Xu, X, Cui, H, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Niemann, G.S, Merkley, E.D, Savchenko, A, Adkins, J.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2012-04-26 | | Release date: | 2012-05-23 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | C-terminal domain of inner membrane protein CigR from Salmonella enterica.
To be Published
|
|
4ELZ
 
 | | CCDBVFI:GYRA14VFI | | Descriptor: | CcdB, DNA gyrase subunit A, GLYCEROL | | Authors: | De Jonge, N, Simic, M, Buts, L, Haesaerts, S, Roelants, K, Garcia-Pino, A, Sterckx, Y, De Greve, H, Lah, J, Loris, R. | | Deposit date: | 2012-04-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Alternative interactions define gyrase specificity in the CcdB family.
Mol.Microbiol., 84, 2012
|
|
4EO4
 
 | | Crystal structure of the yeast mitochondrial threonyl-tRNA synthetase (MST1) in complex with seryl sulfamoyl adenylate | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, Threonine--tRNA ligase, mitochondrial, ... | | Authors: | Peterson, K.M, Ling, J, Simonovic, I, Soll, D, Simonovic, M. | | Deposit date: | 2012-04-13 | | Release date: | 2012-07-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | The mechanism of pre-transfer editing in yeast mitochondrial threonyl-tRNA synthetase.
J.Biol.Chem., 287, 2012
|
|
1UQR
 
 | | Type II 3-dehydroquinate dehydratase (DHQase) from Actinobacillus pleuropneumoniae | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE, SULFATE ION | | Authors: | Maes, D, Gonzalez-Ramirez, L.A, Lopez-Jaramillo, J, Yu, B, De Bondt, H, Zegers, I, Afonina, E, Garcia-Ruiz, J.M, Gulnik, S. | | Deposit date: | 2003-10-16 | | Release date: | 2003-10-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Study of the Type II 3-Dehydroquinate Dehydratase from Actinobacillus Pleuropneumoniae
Acta Crystallogr.,Sect.D, 60, 2004
|
|
4E6T
 
 | | Structure of LpxA from Acinetobacter baumannii at 1.8A resolution (P212121 form) | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, CITRATE ANION | | Authors: | Badger, J, Chie-Leon, B, Logan, C, Sridhar, V, Sankaran, B, Zwart, P.H, Nienaber, V. | | Deposit date: | 2012-03-15 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure determination of LpxA from the lipopolysaccharide-synthesis pathway of Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
1UOF
 
 | | Deacetoxycephalosporin C synthase complexed with Penicillin G | | Descriptor: | DEACETOXYCEPHALOSPORIN C SYNTHETASE, FE (II) ION, PENICILLIN G | | Authors: | Valegard, K, Terwisscha Van scheltinga, A.C, Dubus, A, Oster, L.M, Rhangino, G, Hajdu, J, Andersson, I. | | Deposit date: | 2003-09-16 | | Release date: | 2004-01-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structural Basis of Cephalosporin Formation in a Mononuclear Ferrous Enzyme
Nat.Struct.Mol.Biol., 11, 2004
|
|
4EUK
 
 | | Crystal structure | | Descriptor: | 1,2-ETHANEDIOL, Histidine kinase 5, Histidine-containing phosphotransfer protein 1, ... | | Authors: | Stehle, T, Bauer, J. | | Deposit date: | 2012-04-25 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Function Analysis of Arabidopsis thaliana Histidine Kinase AHK5 Bound to Its Cognate Phosphotransfer Protein AHP1.
Mol Plant, 6, 2013
|
|
4EA6
 
 | | Crystal structure of Fungal lipase from Thermomyces(Humicola) lanuginosa at 2.30 Angstrom resolution. | | Descriptor: | Lipase | | Authors: | Kumar, M, Mukherjee, J, Gupta, M.N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-03-22 | | Release date: | 2012-04-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Fungal lipase from Thermomyces(Humicola) lanuginosa at 2.30 Angstrom resolution.
To be Published
|
|
4EAL
 
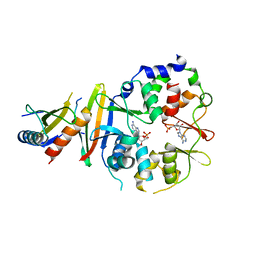 | | Co-crystal of AMPK core with ATP soaked with AMP | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Chen, L, Wang, J, Zhang, Y.-Y, Yan, S.F, Neumann, D, Schlattner, U, Wang, Z.-X, Wu, J.-W. | | Deposit date: | 2012-03-22 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | AMP-activated protein kinase undergoes nucleotide-dependent conformational changes
Nat.Struct.Mol.Biol., 19, 2012
|
|
1UU8
 
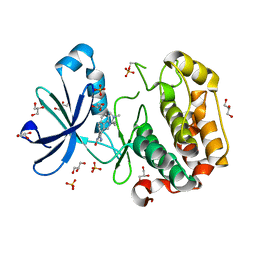 | | Structure of human PDK1 kinase domain in complex with BIM-1 | | Descriptor: | 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE-1, 3-{1-[3-(DIMETHYLAMINO)PROPYL]-1H-INDOL-3-YL}-4-(1H-INDOL-3-YL)-1H-PYRROLE-2,5-DIONE, GLYCEROL, ... | | Authors: | Komander, D, Kular, G.S, Schuttelkopf, A.W, Deak, M, Prakash, K.R, Bain, J, Elliot, M, Garrido-Franco, M, Kozikowski, A.P, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2003-12-16 | | Release date: | 2004-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Interactions of Ly333531 and Other Bisindolyl Maleimide Inhibitors with Pdk1
Structure, 12, 2004
|
|
1V0M
 
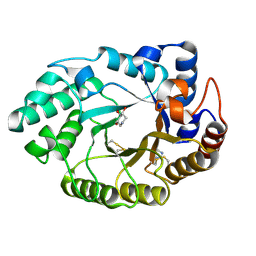 | | Xylanase Xyn10a from Streptomyces lividans in complex with xylobio-deoxynojirimycin at pH 7.5 | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, IMIDAZOLE, PIPERIDINE-3,4,5-TRIOL, ... | | Authors: | Gloster, T.M, Williams, S.J, Roberts, S, Tarling, C.A, Wicki, J, Withers, S.G, Davies, G.J. | | Deposit date: | 2004-03-31 | | Release date: | 2004-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Atomic Resolution Analyses of the Binding of Xylobiose-Derived Deoxynojirimycin and Isofagomine to Xylanase Xyn10A
Chem.Commun.(Camb.), 16, 2004
|
|
4EHF
 
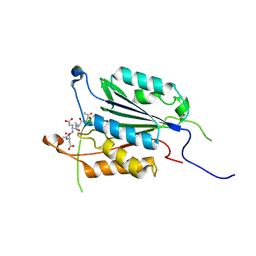 | | Allosteric Modulation of Caspase-3 through Mutagenesis | | Descriptor: | ACE-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE INHIBITOR, Caspase-3 | | Authors: | Walters, J, Schipper, J.L, Swartz, P.D, Mattos, C, Clark, A.C. | | Deposit date: | 2012-04-02 | | Release date: | 2012-06-06 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | Allosteric modulation of caspase 3 through mutagenesis.
Biosci.Rep., 32, 2012
|
|
4EIV
 
 | | 1.37 Angstrom resolution crystal structure of apo-form of a putative deoxyribose-phosphate aldolase from Toxoplasma gondii ME49 | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Deoxyribose-phosphate aldolase | | Authors: | Halavaty, A.S, Ruan, J, Minasov, G, Shuvalova, L, Ueno, A, Igarashi, M, Ngo, H, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-05 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural and Functional Divergence of the Aldolase Fold in Toxoplasma gondii.
J.Mol.Biol., 427, 2015
|
|
1V4P
 
 | |
1V4T
 
 | | Crystal structure of human glucokinase | | Descriptor: | SODIUM ION, SULFATE ION, glucokinase isoform 2 | | Authors: | Kamata, K, Mitsuya, M, Nishimura, T, Eiki, J, Nagata, Y. | | Deposit date: | 2003-11-19 | | Release date: | 2004-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for allosteric regulation of the monomeric allosteric enzyme human glucokinase
Structure, 12, 2004
|
|
4ELK
 
 | | Crystal structure of the Hy19.3 type II NKT TCR | | Descriptor: | ACETATE ION, FORMIC ACID, Hy19.3 TCR alpha chain (mouse variable domain, ... | | Authors: | Girardi, E, Maricic, I, Wang, J, Mac, T.T, Iyer, P, Kumar, V, Zajonc, D.M. | | Deposit date: | 2012-04-10 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Type II natural killer T cells use features of both innate-like and conventional T cells to recognize sulfatide self antigens.
Nat.Immunol., 13, 2012
|
|
