4O42
 
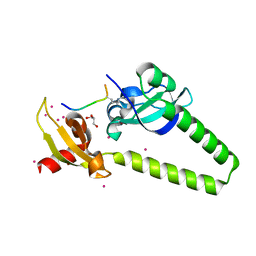 | | Tandem chromodomains of human CHD1 in complex with influenza NS1 C-terminal tail dimethylated at K229 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1, GLYCEROL, Nonstructural protein 1, ... | | Authors: | Qin, S, Xu, C, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural basis for histone mimicry and hijacking of host proteins by influenza virus protein NS1.
Nat Commun, 5, 2014
|
|
4F7N
 
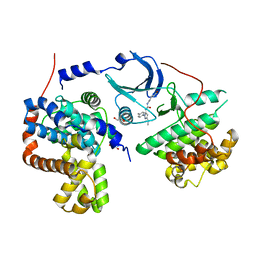 | | Crystal structure of human CDK8/CYCC in complex with compound 11 (1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-(5-hydroxypentyl)urea) | | Descriptor: | 1,2-ETHANEDIOL, 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-(5-hydroxypentyl)urea, Cyclin-C, ... | | Authors: | Schneider, E.V, Boettcher, J, Huber, R, Maskos, K. | | Deposit date: | 2012-05-16 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-kinetic relationship study of CDK8/CycC specific compounds.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4O34
 
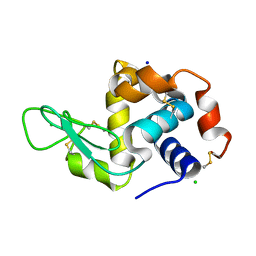 | | Room temperature macromolecular serial crystallography using synchrotron radiation | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Stellato, F, Oberthuer, D, Liang, M, Bean, R, Gati, C, Yefanov, O, Barty, A, Burkhardt, A, Fischer, P, Galli, L, Kirian, R.A, Mayer, J, Pannerselvam, S, Yoon, C.H, Chervinskii, F, Speller, E, White, T.A, Betzel, C, Meents, A, Chapman, H.N. | | Deposit date: | 2013-12-18 | | Release date: | 2014-06-11 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Room-temperature macromolecular serial crystallography using synchrotron radiation.
IUCrJ, 1, 2014
|
|
4O46
 
 | | 14-3-3-gamma in complex with influenza NS1 C-terminal tail phosphorylated at S228 | | Descriptor: | 14-3-3 protein gamma, Nonstructural protein 1, UNKNOWN ATOM OR ION, ... | | Authors: | Qin, S, Liu, Y, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for histone mimicry and hijacking of host proteins by influenza virus protein NS1.
Nat Commun, 5, 2014
|
|
4O94
 
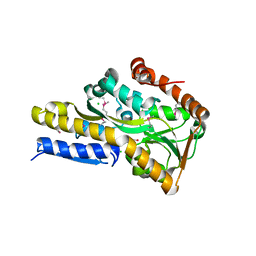 | | Crystal structure of a trap periplasmic solute binding protein from Rhodopseudomonas palustris HaA2 (RPB_3329), Target EFI-510223, with bound succinate | | Descriptor: | CHLORIDE ION, SUCCINIC ACID, TRAP dicarboxylate transporter DctP subunit | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-01-01 | | Release date: | 2014-01-22 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4O7S
 
 | |
4O86
 
 | |
4FLH
 
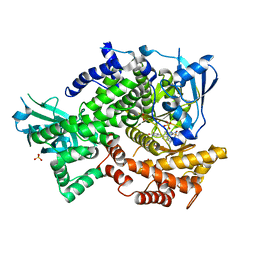 | | Crystal structure of human PI3K-gamma in complex with AMG511 | | Descriptor: | 4-(2-[(5-fluoro-6-methoxypyridin-3-yl)amino]-5-{(1R)-1-[4-(methylsulfonyl)piperazin-1-yl]ethyl}pyridin-3-yl)-6-methyl-1,3,5-triazin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2012-06-14 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Selective Class I Phosphoinositide 3-Kinase Inhibitors: Optimization of a Series of Pyridyltriazines Leading to the Identification of a Clinical Candidate, AMG 511.
J.Med.Chem., 55, 2012
|
|
4FHJ
 
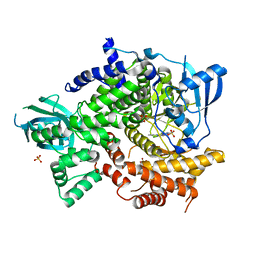 | | Crystal Structure of PI3K-gamma in Complex with Imidazopyridine 2 | | Descriptor: | 3-(4-amino-6-methyl-1,3,5-triazin-2-yl)-N-(1H-pyrazol-5-yl)imidazo[1,2-a]pyridin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Shaffer, P.L, Tang, J, Yakowec, P. | | Deposit date: | 2012-06-06 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery and optimization of potent and selective imidazopyridine and imidazopyridazine mTOR inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4OMO
 
 | | Crystal structure of the c-Src tyrosine kinase SH3 domain mutant Q128E | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NICKEL (II) ION, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Camara-Artigas, A, Bacarizo, J. | | Deposit date: | 2014-01-27 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Electrostatic Effects in the Folding of the SH3 Domain of the c-Src Tyrosine Kinase: pH-Dependence in 3D-Domain Swapping and Amyloid Formation.
Plos One, 9, 2014
|
|
4OUN
 
 | | Crystal Structure of Mini-ribonuclease 3 from Bacillus subtilis | | Descriptor: | Mini-ribonuclease 3 | | Authors: | Chojnowski, G, Czarnecka, J, Nowak, E, Pianka, D, Glow, D, Sabala, I, Skowronek, K, Nowotny, M, Bujnicki, J.M. | | Deposit date: | 2014-02-18 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sequence-specific cleavage of dsRNA by Mini-III RNase
Nucleic Acids Res., 43, 2015
|
|
4OVP
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM SULFITOBACTER sp. NAS-14.1, TARGET EFI-510292, WITH BOUND ALPHA-D-MANURONATE | | Descriptor: | C4-dicarboxylate transport system substrate-binding protein, alpha-D-mannopyranuronic acid | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-11 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4F6S
 
 | | Crystal structure of human CDK8/CYCC in complex with compound 7 (1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]urea) | | Descriptor: | 1,2-ETHANEDIOL, 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]urea, Cyclin-C, ... | | Authors: | Schneider, E.V, Boettcher, J, Huber, R, Maskos, K. | | Deposit date: | 2012-05-15 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-kinetic relationship study of CDK8/CycC specific compounds.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4OVQ
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM ROSEOBACTER DENITRIFICANS, TARGET EFI-510230, WITH BOUND BETA-D-GLUCURONATE | | Descriptor: | CHLORIDE ION, TRAP dicarboxylate ABC transporter, substrate-binding protein, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-11 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4O82
 
 | |
4O8M
 
 | | Crystal structure of a trap periplasmic solute binding protein actinobacillus succinogenes 130z, target EFI-510004, with bound L-galactonate | | Descriptor: | CHLORIDE ION, L-galactonic acid, SULFATE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-28 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4O7M
 
 | | Crystal structure of a trap periplasmic solute binding protein from shewanella loihica PV-4, target EFI-510273, with bound L-malate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, SULFATE ION, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-25 | | Release date: | 2014-03-05 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4O7T
 
 | | SAICAR synthetase (Type-2) in complex with ADP, ASP and TMP | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ASPARTIC ACID, ... | | Authors: | Manjunath, K, Jeyakanthan, J, Sekar, K. | | Deposit date: | 2013-12-26 | | Release date: | 2014-12-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | SAICAR synthetase (Type-2) in complex with ADP, ASP and TMP
To be Published
|
|
8E15
 
 | | A computationally stabilized hMPV F protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F1 protein with Fibritin peptide, F2 protein, ... | | Authors: | Huang, J, Gonzalez, K, Mousa, J, Strauch, E. | | Deposit date: | 2022-08-09 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A general computational design strategy for stabilizing viral class I fusion proteins.
Biorxiv, 2023
|
|
4O7R
 
 | |
4O84
 
 | |
4OQW
 
 | | Crystal structure of mCardinal far-red fluorescent protein | | Descriptor: | Fluorescent protein FP480 | | Authors: | Burg, J.S, Chu, J, Lam, A.J, Lin, M.Z, Garcia, K.C. | | Deposit date: | 2014-02-10 | | Release date: | 2014-03-12 | | Last modified: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Non-invasive intravital imaging of cellular differentiation with a bright red-excitable fluorescent protein.
Nat.Methods, 11, 2014
|
|
4FP7
 
 | |
4OVS
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM SULFUROSPIRILLUM DELEYIANUM DSM 6946 (Sdel_0447), TARGET EFI-510309, WITH BOUND SUCCINATE | | Descriptor: | CHLORIDE ION, SUCCINIC ACID, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-13 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4OML
 
 | |
