1MG5
 
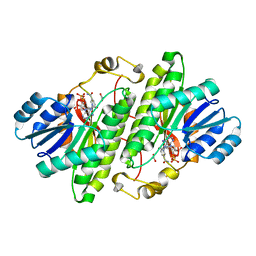 | | Crystal structure of Drosophila melanogaster alcohol dehydrogenase complexed with NADH and acetate at 1.6 A | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ACETATE ION, alcohol dehydrogenase | | Authors: | Benach, J, Atrian, S, Gonzalez-Duarte, R, Ladenstein, R. | | Deposit date: | 2002-08-14 | | Release date: | 2003-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Drosophila alcohol dehydrogenase: acetate-enzyme interactions and novel insights into the effects of electrostatics on catalysis
J.Mol.Biol., 345, 2005
|
|
8UA8
 
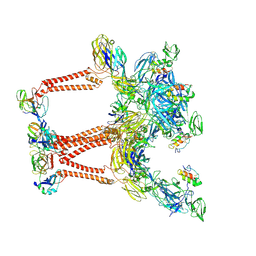 | | Structure of Semliki Forest virus VLP in complex with VLDLR LA2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Assembly protein E3, ... | | Authors: | Abraham, J, Yang, P, Li, W, Fan, X, Pan, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of SFV VLP-VLDLRAD2 complex at the 3-fold axes
To Be Published
|
|
8UA4
 
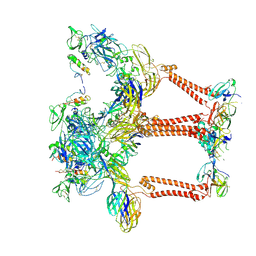 | | Structure of eastern equine encephalitis virus VLP in complex with VLDLR LA1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Capsid protein, ... | | Authors: | Abraham, J, Yang, P, Li, W, Fan, X, Pan, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis for VLDLR recognition by eastern equine encephalitis virus.
Nat Commun, 15, 2024
|
|
8UA9
 
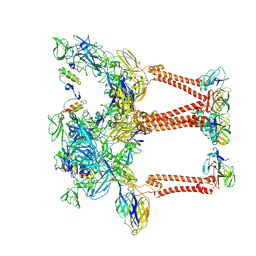 | | Structure of eastern equine encephalitis virus VLP unliganded quasi-threefold spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein, Envelope glycoprotein E1, ... | | Authors: | Abraham, J, Yang, P, Li, W, Fan, X, Pan, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for VLDLR recognition by eastern equine encephalitis virus.
Nat Commun, 15, 2024
|
|
1W0Y
 
 | | tf7a_3771 complex | | Descriptor: | 4-(4-BENZYLOXY-2-METHANESULFONYLAMINO-5-METHOXY-BENZYLAMINO)-BENZAMIDINE, BLOOD COAGULATION FACTOR VIIA, CACODYLATE ION, ... | | Authors: | Banner, D.W, D'Arcy, A, Groebke-Zbinden, K, Ackermann, J, Kirchhofer, D, Ji, Y.-H, Tschopp, T.B, Wallbaum, S, Weber, L. | | Deposit date: | 2004-05-27 | | Release date: | 2005-01-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design of Selective Phenylglycine Amide Tissue Factor/Factor Viia Inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1W9Q
 
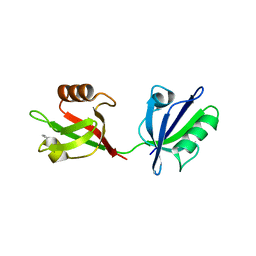 | | Crystal structure of the PDZ tandem of human syntenin in complex with TNEFAF peptide | | Descriptor: | BENZOIC ACID, SYNTENIN 1, TNEFAF PEPTIDE | | Authors: | Grembecka, J, Cierpicki, T, Devedjiev, Y, Cooper, D.R, Derewenda, Z.S. | | Deposit date: | 2004-10-15 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Binding of the Pdz Tandem of Syntenin to Target Proteins.
Biochemistry, 45, 2006
|
|
1VD6
 
 | |
4US1
 
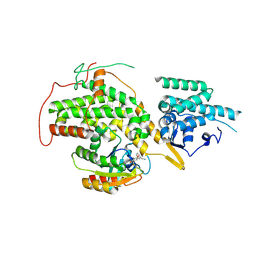 | | The crystal structure of H-Ras and SOS in complex with ligands | | Descriptor: | (3S)-3-[3-(aminomethyl)phenyl]-1-ethylpyrrolidine-2,5-dione, GTPASE HRAS, SON OF SEVENLESS HOMOLOG 1 | | Authors: | Winter, J.J.G, Anderson, M, Blades, K, Brassington, C, Breeze, A.L, Chresta, C, Embrey, K, Fairley, G, Faulder, P, Finlay, M.R.V, Kettle, J.G, Nowak, T, Overman, R, Patel, S.J, Perkins, P, Spadola, L, Tart, J, Tucker, J, Wrigley, G. | | Deposit date: | 2014-07-02 | | Release date: | 2015-03-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Small Molecule Binding Sites on the Ras:SOS Complex Can be Exploited for Inhibition of Ras Activation.
J.Med.Chem., 58, 2015
|
|
1VJY
 
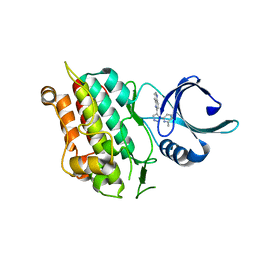 | | Crystal Structure of a Naphthyridine Inhibitor of Human TGF-beta Type I Receptor | | Descriptor: | 2-[5-(6-METHYLPYRIDIN-2-YL)-2,3-DIHYDRO-1H-PYRAZOL-4-YL]-1,5-NAPHTHYRIDINE, TGF-beta receptor type I | | Authors: | Gellibert, F, Woolven, J, Fouchet, M.-H, Mathews, N, Goodland, H, Lovegrove, V, Laroze, A, Nguyen, V.-L, Sautet, S, Wang, R, Janson, C, Smith, W, Krysa, G, Boullay, V, de Gouville, A.-C, Huet, S, Hartley, D. | | Deposit date: | 2004-04-07 | | Release date: | 2004-08-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of 1,5-Naphthyridine Derivatives as a Novel Series of Potent and Selective TGF-beta Type I Receptor Inhibitors.
J.Med.Chem., 47, 2004
|
|
9PAP
 
 | |
4V4W
 
 | |
1VKJ
 
 | | Crystal structure of heparan sulfate 3-O-sulfotransferase isoform 1 in the presence of PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, SULFATE ION, heparan sulfate (glucosamine) 3-O-sulfotransferase 1 | | Authors: | Thorp, S, Lee, K.A, Negishi, M, Linhardt, R.J, Liu, J, Pedersen, L.C. | | Deposit date: | 2004-05-25 | | Release date: | 2004-06-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and mutational analysis of heparan sulfate 3-O-sulfotransferase isoform 1
J.Biol.Chem., 279, 2004
|
|
4V4V
 
 | |
4URW
 
 | | The crystal structure of H-Ras and SOS in complex with ligands | | Descriptor: | 2-(2,6-DIMETHYLPHENYL)-4-(METHYLSULFANYL)-6-(PIPERAZIN-1-YL)-1,3,5-TRIAZINE, GTPASE HRAS, SON OF SEVENLESS HOMOLOG 1 | | Authors: | Winter, J.J.G, Anderson, M, Blades, K, Brassington, C, Breeze, A.L, Chresta, C, Embrey, K, Fairley, G, Faulder, P, Finlay, M.R.V, Kettle, J.G, Nowak, T, Overman, R, Patel, S.J, Perkins, P, Spadola, L, Tart, J, Tucker, J, Wrigley, G. | | Deposit date: | 2014-07-02 | | Release date: | 2015-03-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Small Molecule Binding Sites on the Ras:SOS Complex Can be Exploited for Inhibition of Ras Activation.
J.Med.Chem., 58, 2015
|
|
1W5W
 
 | | HIV-1 protease in complex with fluoro substituted diol-based C2- symmetric inhibitor | | Descriptor: | (2R,3R,4R,5R)-2,5-BIS[(2,4-DIFLUOROBENZYL)OXY]-3,4-DIHYDROXY-N,N'-BIS[(1R,2S)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL]HEXAN EDIAMIDE, POL POLYPROTEIN | | Authors: | Lindberg, J, Pyring, D, Loewgren, S, Rosenquist, A, Zuccarello, G, Kvarnstroem, I, Zhang, H, Vrang, L, Claesson, B, Hallberg, A, Samuelsson, B, Unge, T. | | Deposit date: | 2004-08-10 | | Release date: | 2004-12-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Symmetric Fluoro-Substituted Diol-Based HIV Protease Inhibitors. Ortho-Fluorinated and Meta-Fluorinated P1/P1'-Benzyloxy Side Groups Significantly Improve the Antiviral Activity and Preserve Binding Efficacy
Eur.J.Biochem., 271, 2004
|
|
4URY
 
 | | The crystal structure of H-Ras and SOS in complex with ligands | | Descriptor: | GTPASE HRAS, N-[(4-aminophenyl)sulfonyl]cyclopropanecarboxamide, SON OF SEVENLESS HOMOLOG 1 | | Authors: | Winter, J.J.G, Anderson, M, Blades, K, Brassington, C, Breeze, A.L, Chresta, C, Embrey, K, Fairley, G, Faulder, P, Finlay, M.R.V, Kettle, J.G, Nowak, T, Overman, R, Patel, S.J, Perkins, P, Spadola, L, Tart, J, Tucker, J, Wrigley, G. | | Deposit date: | 2014-07-02 | | Release date: | 2015-03-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Small Molecule Binding Sites on the Ras:SOS Complex Can be Exploited for Inhibition of Ras Activation.
J.Med.Chem., 58, 2015
|
|
1VJA
 
 | | Urokinase Plasminogen Activator B-Chain-JT463 Complex | | Descriptor: | N-(BENZYLSULFONYL)SERYL-N~1~-{4-[(Z)-AMINO(IMINO)METHYL]BENZYL}SERINAMIDE, SULFATE ION, plasminogen activator, ... | | Authors: | Schweinitz, A, Steinmetzer, T, Banke, I.J, Arlt, M.J.E, Stuerzebecher, A, Schuster, O, Geissler, A, Giersiefen, H, Zeslawska, E, Jacob, U, Kruger, A, Stuerzebecher, J. | | Deposit date: | 2004-02-03 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of novel and selective inhibitors of urokinase-type plasminogen activator with improved pharmacokinetic properties for use as antimetastatic agents
J.Biol.Chem., 279, 2004
|
|
1VR2
 
 | | HUMAN VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR 2 (KDR) KINASE DOMAIN | | Descriptor: | PROTEIN (VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR KINASE) | | Authors: | Mctigue, M, Wickersham, J, Pinko, C, Showalter, R, Parast, C, Tempczyk-Russell, A, Gehring, M, Mroczkowski, B, Kan, C, Villafranca, J, Appelt, K. | | Deposit date: | 1998-12-03 | | Release date: | 2000-03-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the kinase domain of human vascular endothelial growth factor receptor 2: a key enzyme in angiogenesis.
Structure Fold.Des., 7, 1999
|
|
1VRX
 
 | | Endocellulase e1 from acidothermus cellulolyticus mutant y245g | | Descriptor: | ENDOCELLULASE E1 FROM A. CELLULOLYTICUS | | Authors: | Baker, J.O, McCarley, J.R, Lovett, R, Yu, C.H, Adney, W.S, Rignall, T.R, Vinzant, T.B, Decker, S.R, Sakon, J, Himmel, M.E. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catalytically enhanced endocellulase Cel5A from Acidothermus cellulolyticus.
Appl.Biochem.Biotechnol., 121-124, 2005
|
|
1VKG
 
 | | Crystal Structure of Human HDAC8 complexed with CRA-19156 | | Descriptor: | 5-(4-METHYL-BENZOYLAMINO)-BIPHENYL-3,4'-DICARBOXYLIC ACID 3-DIMETHYLAMIDE-4'-HYDROXYAMIDE, Histone deacetylase 8, SODIUM ION, ... | | Authors: | Somoza, J.R, Skene, R.J, Katz, B.A, Mol, C, Ho, J.D, Jennings, A.J, Luong, C, Arvai, A, Buggy, J.J, Chi, E, Tang, J, Sang, B.-C, Verner, E, Wynands, R, Leahy, E.M, Dougan, D.R, Snell, G, Navre, M, Knuth, M.W, Swanson, R.V, McRee, D.E, Tari, L.W. | | Deposit date: | 2004-05-13 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Snapshots of Human HDAC8 Provide Insights into the Class I Histone Deacetylases
Structure, 12, 2004
|
|
1VS0
 
 | | Crystal Structure of the Ligase Domain from M. tuberculosis LigD at 2.4A | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Putative DNA ligase-like protein Rv0938/MT0965, ... | | Authors: | Akey, D, Martins, A, Aniukwu, J, Glickman, M.S, Shuman, S, Berger, J.M, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-01-27 | | Release date: | 2006-02-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure and Nonhomologous End-joining Function of the Ligase Component of Mycobacterium DNA Ligase D.
J.Biol.Chem., 281, 2006
|
|
5LWJ
 
 | | Solution NMR structure of the GTP binding Class II RNA aptamer-ligand-complex containing a protonated adenine nucleotide with a highly shifted pKa. | | Descriptor: | GTP Class II RNA (34-MER), GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Wolter, A.C, Weickhmann, A.K, Nasiri, A.H, Hantke, K, Ohlenschlaeger, O, Wunderlich, C.H, Kreutz, C, Duchardt-Ferner, E, Woehnert, J. | | Deposit date: | 2016-09-17 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Stably Protonated Adenine Nucleotide with a Highly Shifted pKa Value Stabilizes the Tertiary Structure of a GTP-Binding RNA Aptamer.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
1VTJ
 
 | | MOLECULAR STRUCTURE OF THE NETROPSIN-D(CGCGATATCGCG) COMPLEX: DNA CONFORMATION IN AN ALTERNATING AT SEGMENT; CONFORMATION 1 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*TP*AP*TP*CP*GP*CP*G)-3'), NETROPSIN | | Authors: | Coll, M, Aymami, J, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J. | | Deposit date: | 1999-09-14 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Structure of the Netropsin-d(CGCGATATCGCG) Complex: DNA Conformation in an Alternating AT Segment
Biochemistry, 28, 1989
|
|
1VPD
 
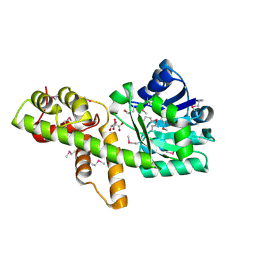 | | X-Ray Crystal Structure of Tartronate Semialdehyde Reductase [Salmonella Typhimurium LT2] | | Descriptor: | CHLORIDE ION, L(+)-TARTARIC ACID, TARTRONATE SEMIALDEHYDE REDUCTASE | | Authors: | Osipiuk, J, Zhou, M, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-10-22 | | Release date: | 2004-10-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray crystal structure of GarR-tartronate semialdehyde reductase from Salmonella typhimurium.
J Struct Funct Genomics, 10, 2009
|
|
1VZY
 
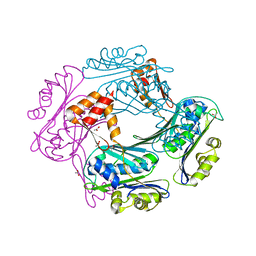 | | Crystal structure of the Bacillus subtilis HSP33 | | Descriptor: | 33 KDA CHAPERONIN, ACETATE ION, ZINC ION | | Authors: | Janda, I.K, Devedjiev, Y, Derewenda, U, Dauter, Z, Bielnicki, J, Cooper, D.R, Joachimiak, A, Derewenda, Z.S, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-29 | | Release date: | 2004-10-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The crystal structure of the reduced, Zn2+-bound form of the B. subtilis Hsp33 chaperone and its implications for the activation mechanism.
Structure, 12, 2004
|
|
