2KE8
 
 | | NMR solution structure of metal-modified DNA | | Descriptor: | DNA (5'-D(*TP*TP*AP*AP*TP*TP*TP*(D33)P*(D33)P*(D33)P*AP*AP*AP*TP*TP*AP*A)-3'), SILVER ION | | Authors: | Johannsen, S, Duepre, N, Boehme, D, Mueller, J, Sigel, R.K.O. | | Deposit date: | 2009-01-27 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA double helix with consecutive metal-mediated base pairs.
Nat.Chem., 2, 2010
|
|
2KPU
 
 | | NMR Structure of YbbR family protein Dhaf_0833 (residues 32-118) from Desulfitobacterium hafniense DCB-2: Northeast Structural Genomics Consortium target DhR29B | | Descriptor: | YbbR family protein | | Authors: | Cort, J.R, Ramelot, T.A, Yang, Y, Belote, R.L, Ciccosanti, C, Haleema, J, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structures of domains I and IV from YbbR are representative of a widely distributed protein family.
Protein Sci., 20, 2011
|
|
1H9Y
 
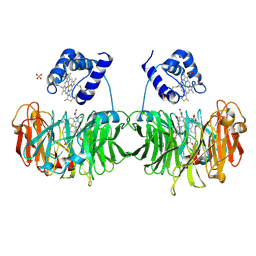 | |
2KPH
 
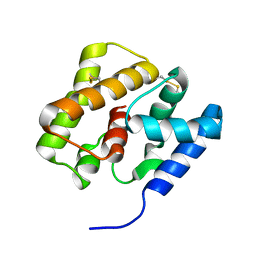 | | NMR Structure of AtraPBP1 at pH 4.5 | | Descriptor: | Pheromone binding protein | | Authors: | Ames, J. | | Deposit date: | 2009-10-15 | | Release date: | 2010-02-02 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Navel Orangeworm Moth Pheromone-Binding Protein (AtraPBP1): Implications for pH-Sensitive Pheromone Detection .
Biochemistry, 49, 2010
|
|
1H8D
 
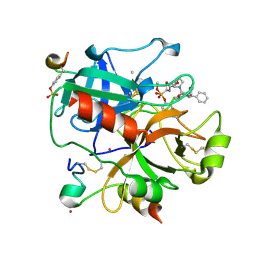 | | X-ray structure of the human alpha-thrombin complex with a tripeptide phosphonate inhibitor. | | Descriptor: | HIRUDIN I, N-[(benzyloxy)carbonyl]-beta-phenyl-D-phenylalanyl-N-{(1S,3E)-1-[dihydroxy(diphenoxy)-lambda~5~-phosphanyl]-4-methoxybut-3-en-1-yl}-L-prolinamide, THROMBIN | | Authors: | Skordalakes, E, Dodson, G.G, Green, D, Deadman, J. | | Deposit date: | 2001-02-01 | | Release date: | 2001-02-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibition of Human Alpha-Thrombin by a Phosphonate Tripeptide Proceeds Via a Metastable Pentacoordinated Phosphorus Intermediate
J.Mol.Biol., 311, 2001
|
|
1HAI
 
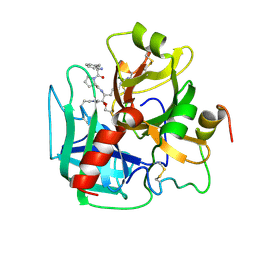 | | THE ISOMORPHOUS STRUCTURES OF PRETHROMBIN2, HIRUGEN-AND PPACK-THROMBIN: CHANGES ACCOMPANYING ACTIVATION AND EXOSITE BINDING TO THROMBIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), ... | | Authors: | Tulinsky, A, Vijayalakshmi, J. | | Deposit date: | 1994-06-27 | | Release date: | 1994-12-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The isomorphous structures of prethrombin2, hirugen-, and PPACK-thrombin: changes accompanying activation and exosite binding to thrombin.
Protein Sci., 3, 1994
|
|
2HT9
 
 | | The structure of dimeric human glutaredoxin 2 | | Descriptor: | 12-mer peptide, FE2/S2 (INORGANIC) CLUSTER, GLUTATHIONE, ... | | Authors: | Johansson, C, Smee, C, Kavanagh, K.L, Debreczeni, J, von Delft, F, Gileadi, O, Arrowsmith, C, Weigelt, J, Edwards, A, Sundstrom, M, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-07-25 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reversible sequestration of active site cysteines in a 2Fe-2S-bridged dimer provides a mechanism for glutaredoxin 2 regulation in human mitochondria
J.Biol.Chem., 282, 2007
|
|
1H9X
 
 | | Cytochrome cd1 Nitrite Reductase, reduced form | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CYTOCHROME CD1 NITRITE REDUCTASE, HEME C, ... | | Authors: | Sjogren, T, Hajdu, J. | | Deposit date: | 2001-03-23 | | Release date: | 2001-08-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of an Alternative Form of Paracoccus Pantotrophus Cytochrome Cd1 Nitrite Reductase
J.Biol.Chem., 276, 2001
|
|
1GYO
 
 | | Crystal structure of the di-tetraheme cytochrome c3 from Desulfovibrio gigas at 1.2 Angstrom resolution | | Descriptor: | CYTOCHROME C3, A DIMERIC CLASS III C-TYPE CYTOCHROME, GLYCEROL, ... | | Authors: | Aragao, D, Frazao, C, Sieker, L, Sheldrick, G.M, Legall, J, Carrondo, M.A. | | Deposit date: | 2002-04-29 | | Release date: | 2002-05-24 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of Dimeric Cytochrome C3 from Desulfovibrio Gigas at 1.2 A Resolution
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2I7Q
 
 | | Crystal structure of Human Choline Kinase A | | Descriptor: | CHLORIDE ION, Choline kinase alpha, UNKNOWN ATOM OR ION | | Authors: | Rabeh, W.M, Tempel, W, Nedyalkova, L, Wasney, G, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-31 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Human Choline Kinase A
To be Published
|
|
2GFT
 
 | | Crystal structure of the E263A nucleophile mutant of Bacillus licheniformis endo-beta-1,4-galactanase in complex with galactotriose | | Descriptor: | CALCIUM ION, Glycosyl Hydrolase Family 53, beta-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose | | Authors: | Welner, D, Le Nours, J, De Maria, L, Jorgensen, C.T, Christensen, L.L.H, Larsen, S, Lo Leggio, L. | | Deposit date: | 2006-03-23 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Oligosaccharide binding of Bacillus licheniformis endo-beta-1,4-galactanase
To be Published
|
|
2GDC
 
 | | Structure of Vinculin VD1 / IpaA560-633 complex | | Descriptor: | Invasin ipaA, Vinculin | | Authors: | Hamiaux, C, van Eerde, A, Parsot, C, Broos, J, Dijkstra, B.W. | | Deposit date: | 2006-03-15 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural mimicry for vinculin activation by IpaA, a virulence factor of Shigella flexneri.
Embo Rep., 7, 2006
|
|
2KTX
 
 | | COMPLETE KALIOTOXIN FROM ANDROCTONUS MAURETANICUS MAURETANICUS, NMR, 18 STRUCTURES | | Descriptor: | KALIOTOXIN | | Authors: | Gairi, M, Romi, R, Fernandez, I, Rochat, H, Martin-Eauclaire, M.-F, Van Rietschtoten, J, Pons, M, Giralt, E. | | Deposit date: | 1997-02-27 | | Release date: | 1997-06-16 | | Last modified: | 2017-12-20 | | Method: | SOLUTION NMR | | Cite: | 3D structure of kaliotoxin: is residue 34 a key for channel selectivity?
J.Pept.Sci., 3, 1997
|
|
2K7P
 
 | | Filamin A Ig-like domains 16-17 | | Descriptor: | Filamin-A | | Authors: | Heikkinen, O.K, Kilpelainen, I, Koskela, H, Permi, P, Heikkinen, S, Ylanne, J. | | Deposit date: | 2008-08-19 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Atomic structures of two novel immunoglobulin-like domain pairs in the actin cross-linking protein filamin
J.Biol.Chem., 284, 2009
|
|
3LMN
 
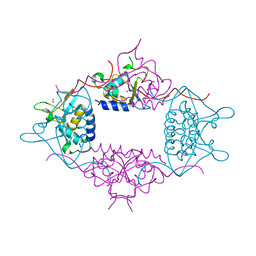 | | Oligomeric structure of the DUSP domain of human USP15 | | Descriptor: | ACETIC ACID, FORMIC ACID, Ubiquitin carboxyl-terminal hydrolase 15 | | Authors: | Walker, J.R, Asinas, A, Avvakumov, G.V, Alenkin, D, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Dhe-Paganon, S. | | Deposit date: | 2010-01-31 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Human Ubiquitin-Specific Protease 15 DUSP Domain
To be Published
|
|
3PI2
 
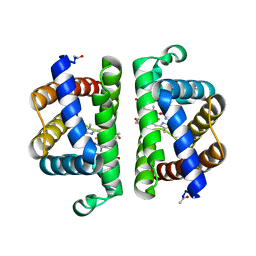 | | Crystallographic Structure of HbII-oxy from Lucina pectinata at pH 8.0 | | Descriptor: | FORMIC ACID, Hemoglobin II, OXYGEN MOLECULE, ... | | Authors: | Gavira, J.A, Nieves-Marrero, C.A, Ruiz-Martinez, C.R, Estremera-Andujar, R.A, Lopez-Garriga, J, Garcia-Ruiz, J.M. | | Deposit date: | 2010-11-05 | | Release date: | 2011-11-09 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | pH-dependence crystallographic studies of the oxygen carrier hemoglobin II from Lucina pectinata
To be Published
|
|
3LN6
 
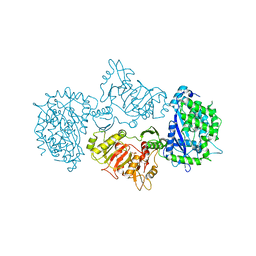 | |
3LR7
 
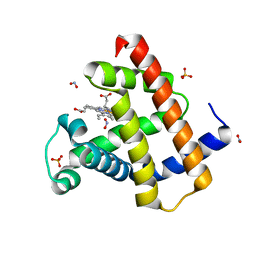 | | Ferric horse heart myoglobin, nitrite adduct | | Descriptor: | Myoglobin, NITRITE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Yi, J, Richter-Addo, G.B. | | Deposit date: | 2010-02-10 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Synchrotron X-ray-Induced Photoreduction of Ferric Myoglobin Nitrite Crystals Gives the Ferrous Derivative with Retention of the O-Bonded Nitrite Ligand.
Biochemistry, 49, 2010
|
|
4FYP
 
 | | Crystal Structure of Plant Vegetative Storage Protein | | Descriptor: | MAGNESIUM ION, Vegetative storage protein 1 | | Authors: | Chen, Y, Wei, J, Wang, M, Gong, W, Zhang, M. | | Deposit date: | 2012-07-05 | | Release date: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of Arabidopsis VSP1 reveals the plant class C-like phosphatase structure of the DDDD superfamily of phosphohydrolases
Plos One, 7, 2012
|
|
3LS7
 
 | | Crystal structure of Thermolysin in complex with Xenon | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Behnen, J, Klebe, G, Heine, A, Brumshtein, B. | | Deposit date: | 2010-02-12 | | Release date: | 2011-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Old acquantance rediscovered, use of xenon/protein complexes as a generic tool for SAD phasing of in house data
To be Published
|
|
4G1M
 
 | | Re-refinement of alpha V beta 3 structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Springer, T.A, Mi, L, Zhu, J. | | Deposit date: | 2012-07-10 | | Release date: | 2012-12-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Alpha V Beta 3 Integrin Crystal Structures and their Functional Implications
Biochemistry, 51, 2012
|
|
3L6R
 
 | | The structure of mammalian serine racemase: Evidence for conformational changes upon inhibitor binding | | Descriptor: | MALONATE ION, MANGANESE (II) ION, Serine racemase | | Authors: | Smith, M.A, Mack, V, Ebneth, A, Cesura, A, Felicetti, B, Barker, J. | | Deposit date: | 2009-12-24 | | Release date: | 2010-01-26 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of mammalian serine racemase: evidence for conformational changes upon inhibitor binding.
J.Biol.Chem., 285, 2010
|
|
3LDA
 
 | | Yeast Rad51 H352Y Filament Interface Mutant | | Descriptor: | CHLORIDE ION, DNA repair protein RAD51 | | Authors: | Villanueva, N.L, Chen, J, Morrical, S.W, Rould, M.A. | | Deposit date: | 2010-01-12 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the mechanism of Rad51 recombinase from the structure and properties of a filament interface mutant.
Nucleic Acids Res., 38, 2010
|
|
4G6P
 
 | | Minimal Hairpin Ribozyme in the Precatalytic State with A38P Variation | | Descriptor: | COBALT HEXAMMINE(III), Loop A Substrate strand, Loop A and Loop B Ribozyme strand, ... | | Authors: | Liberman, J.A, Jenkins, J.L, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2012-07-19 | | Release date: | 2012-08-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.641 Å) | | Cite: | A Transition-State Interaction Shifts Nucleobase Ionization toward Neutrality To Facilitate Small Ribozyme Catalysis.
J.Am.Chem.Soc., 134, 2012
|
|
4G6S
 
 | | Minimal Hairpin Ribozyme in the Transition State with A38P Variation | | Descriptor: | COBALT HEXAMMINE(III), Loop A Substrate strand, Loop A and Loop B Ribozyme strand, ... | | Authors: | Liberman, J.A, Jenkins, J.L, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2012-07-19 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | A Transition-State Interaction Shifts Nucleobase Ionization toward Neutrality To Facilitate Small Ribozyme Catalysis.
J.Am.Chem.Soc., 134, 2012
|
|
