4A5X
 
 | | Structures of MITD1 | | Descriptor: | 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, CHARGED MULTIVESICULAR BODY PROTEIN 1A, GLYCEROL, ... | | Authors: | Hadders, M.A, Agromayor, M, Caballe, A, Obita, T, Perisic, O, Williams, R.L, Martin-Serrano, J. | | Deposit date: | 2011-10-28 | | Release date: | 2012-11-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Escrt-III Binding Protein Mitd1 is Involved in Cytokinesis and Has an Unanticipated Pld Fold that Binds Membranes.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1KSQ
 
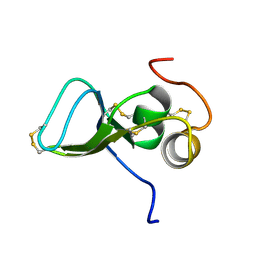 | | NMR Study of the Third TB Domain from Latent Transforming Growth Factor-beta Binding Protein-1 | | Descriptor: | LATENT TRANSFORMING GROWTH FACTOR BETA BINDING PROTEIN 1 | | Authors: | Lack, J, O'leary, J.M, Knott, V, Yuan, X, Rifkin, D.B, Handford, P.A, Downing, A.K. | | Deposit date: | 2002-01-14 | | Release date: | 2003-08-26 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Third TB Domain from LTBP1 Provides Insight into Assembly
of the Large Latent Complex that Sequesters Latent TGF-beta.
J.Mol.Biol., 334, 2003
|
|
6HT5
 
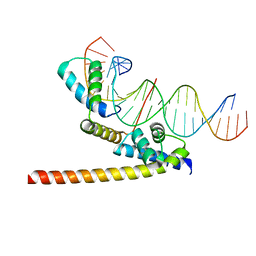 | | Oct4/Sox2:UTF1 structure | | Descriptor: | DNA (5'-D(*TP*TP*CP*AP*CP*TP*AP*GP*CP*AP*TP*AP*AP*CP*AP*AP*TP*GP*AP*GP*G)-3'), DNA (5'-D(P*CP*CP*TP*CP*AP*TP*TP*GP*TP*TP*AP*TP*GP*CP*TP*AP*GP*TP*GP*AP*AP*G)-3'), POU domain, ... | | Authors: | Vahokoski, J, Meusch, D, Groves, M, Pogenberg, V, Wilmanns, M. | | Deposit date: | 2018-10-03 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.451 Å) | | Cite: | Oct4/Sox2:UTF1 structure
To Be Published
|
|
4AME
 
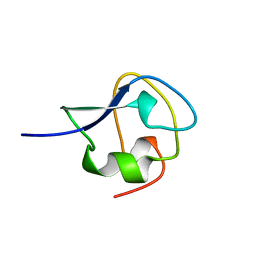 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T18A | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
3D1G
 
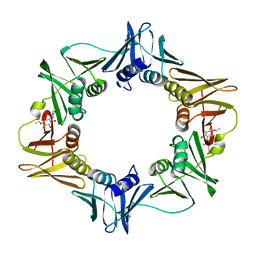 | | Structure of a small molecule inhibitor bound to a DNA sliding clamp | | Descriptor: | DNA polymerase III subunit beta, [(5R)-5-(2,3-dibromo-5-ethoxy-4-hydroxybenzyl)-4-oxo-2-thioxo-1,3-thiazolidin-3-yl]acetic acid | | Authors: | Georgescu, R.E, Yurieva, O, Seung-Sup, K, Kuriyan, J, Kong, X.-P, O'Donnell, M. | | Deposit date: | 2008-05-05 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of a small-molecule inhibitor of a DNA polymerase sliding clamp.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4AE9
 
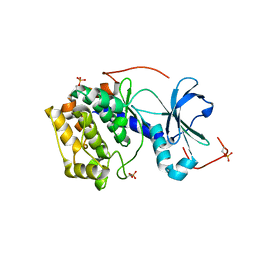 | | Structure and function of the Human Sperm-Specific Isoform of Protein Kinase A (PKA) Catalytic Subunit C alpha 2 | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA | | Authors: | Hereng, T.H, Backe, P.H, Kahmann, J, Scheich, C, Bjoras, M, Skalhegg, B.S, Rosendal, K.R. | | Deposit date: | 2012-01-09 | | Release date: | 2012-04-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Function of the Human Sperm-Specific Isoform of Protein Kinase a (Pka) Catalytic Subunit Calpha2
J.Struct.Biol., 178, 2012
|
|
4AFV
 
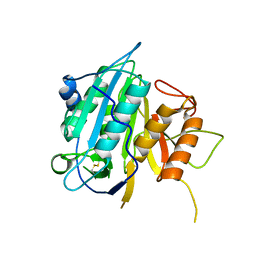 | | THE STRUCTURE OF METACASPASE 2 FROM T. BRUCEI DETERMINED IN THE PRESENCE OF CALCIUM CHLORIDE | | Descriptor: | METACASPASE MCA2 | | Authors: | McLuskey, K, Rudolf, J, Isaacs, N.W, Coombs, G.H, Moss, C.X, Mottram, J.C. | | Deposit date: | 2012-01-23 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of a Trypanosoma Brucei Metacaspase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3D29
 
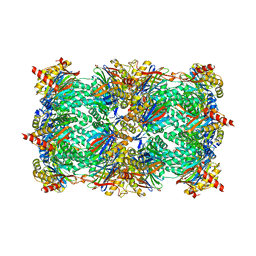 | | Proteasome Inhibition by Fellutamide B | | Descriptor: | (3R)-3-HYDROXYDODECANOIC ACID, Fellutamide B, PRE10 isoform 1, ... | | Authors: | Groll, M, Hines, J, Fahnestock, M, Crews, M.C. | | Deposit date: | 2008-05-07 | | Release date: | 2008-06-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Proteasome Inhibition by Fellutamide B Induces Nerve Growth Factor Synthesis
Chem.Biol., 15, 2008
|
|
6CPH
 
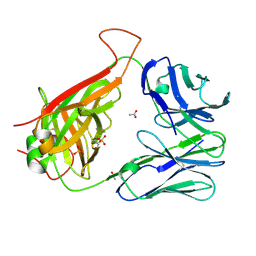 | | Crystal structure of the F24 TCR | | Descriptor: | ACETATE ION, SULFATE ION, TCR alpha chain, ... | | Authors: | Farenc, C, Gras, S, Rossjohn, J. | | Deposit date: | 2018-03-13 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | CD4+T cell-mediated HLA class II cross-restriction in HIV controllers.
Sci Immunol, 3, 2018
|
|
4A7I
 
 | | Factor Xa in complex with a potent 2-amino-ethane sulfonamide inhibitor | | Descriptor: | 5-CHLORO-THIOPHENE-2-CARBOXYLIC ACID [2-(1--ISOPROPYL-PIPERIDIN-4-YLSULFAMOYL)-ETHYL]-AMIDE, ACTIVATED FACTOR XA HEAVY CHAIN XA, CALCIUM ION, ... | | Authors: | Nazare, M, Matter, H, Will, D.W, Wagner, M, Urmann, M, Czech, J, Schreuder, H, Bauer, A, Ritter, K, Wehner, V. | | Deposit date: | 2011-11-14 | | Release date: | 2012-02-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment Deconstruction of Small, Potent Factor Xa Inhibitors: Exploring the Superadditivity Energetics of Fragment Linking in Protein-Ligand Complexes.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
5VAC
 
 | | Crystal Structure of ATXR5 SET domain in complex with K36me3 histone H3 peptide | | Descriptor: | DIMETHYL SULFOXIDE, Histone H3.2, Probable Histone-lysine N-methyltransferase ATXR5, ... | | Authors: | Bergamin, E, Sarvan, S, Malette, J, Eram, M, Yeung, S, Mongeon, V, Joshi, M, Brunzelle, J.S, Michaels, S.D, Blais, A, Vedadi, M, Couture, J.F. | | Deposit date: | 2017-03-24 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Molecular basis for the methylation specificity of ATXR5 for histone H3.
Nucleic Acids Res., 45, 2017
|
|
6CS0
 
 | | SARS Spike Glycoprotein, Trypsin-cleaved, Stabilized variant, one S1 CTD in an upwards conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
3D65
 
 | | Crystal structure of Textilinin-1, a Kunitz-type serine protease inhibitor from the Australian Common Brown snake venom, in complex with trypsin | | Descriptor: | CALCIUM ION, Cationic trypsin, Textilinin | | Authors: | Millers, E.-K.I, Masci, P.P, Lavin, M.F, de Jersey, J, Guddat, L.W. | | Deposit date: | 2008-05-19 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of Textilinin-1, a Kunitz-type serine protease inhibitor from the Australian Common Brown snake venom, in complex with trypsin
To be Published
|
|
4ANB
 
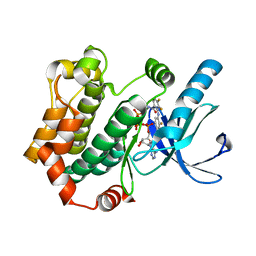 | | Crystal structures of human MEK1 with carboxamide-based allosteric inhibitor XL518 (GDC-0973), or related analogs. | | Descriptor: | DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Rice, K.D, Aay, N, Anand, N.K, Blazey, C.M, Bowles, O.J, Bussenius, J, Costanzo, S, Curtis, J.K, Defina, S.C, Dubenko, L, Engst, S, Joshi, A.A, Kennedy, A.R, Kim, A.I, Koltun, E.S, Lougheed, J.C, Manalo, J.C.L, Martini, J.F, Nuss, J.M, Peto, C.J, Tsang, T.H, Yu, P, Johnston, S. | | Deposit date: | 2012-03-16 | | Release date: | 2012-12-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel Carboxamide-Based Allosteric Mek Inhibitors: Discovery and Optimization Efforts Toward Xl518 (Gdc-0973)
Acs Med.Chem.Lett., 3, 2012
|
|
7T0L
 
 | |
6CT0
 
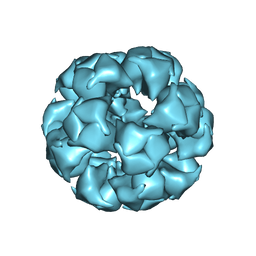 | | Atomic Structure of the E2 Inner Core of Human Pyruvate Dehydrogenase Complex | | Descriptor: | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial | | Authors: | Jiang, J, Baiesc, F.L, Hiromasa, Y, Yu, X, Hui, W.H, Dai, X, Roche, T.E, Zhou, Z.H. | | Deposit date: | 2018-03-21 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Atomic Structure of the E2 Inner Core of Human Pyruvate Dehydrogenase Complex.
Biochemistry, 57, 2018
|
|
3D6W
 
 | | LytTr DNA-binding domain of putative methyl-accepting/DNA response regulator from Bacillus cereus. | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-20 | | Release date: | 2008-07-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystal structure of LytTr DNA-binding domain of putative methyl-accepting/DNA response regulator from Bacillus cereus.
To be Published
|
|
4ANF
 
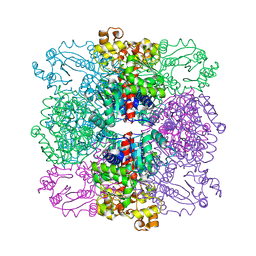 | | Structure of the ornithine carbamoyltransferase from Mycoplasma penetrans with a P23 Space group | | Descriptor: | ORNITHINE CARBAMOYLTRANSFERASE, CATABOLIC | | Authors: | Gallego, P, Benach, J, Planell, R, Querol, E, Perez-Pons, J.A, Reverter, D. | | Deposit date: | 2012-03-16 | | Release date: | 2012-03-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Characterization of the Enzymes Composing the Arginine Deiminase Pathway in Mycoplasma Penetrans.
Plos One, 7, 2012
|
|
3D7F
 
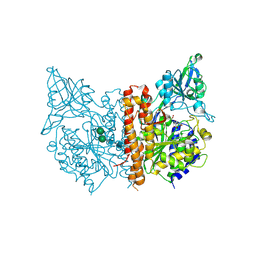 | |
3IC5
 
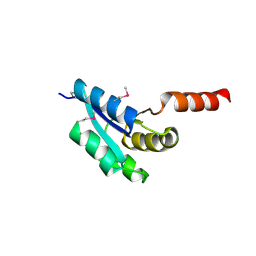 | |
6CB9
 
 | | Segment AALQSS from the low complexity domain of TDP-43, residues 328-333 | | Descriptor: | AALQSS | | Authors: | Guenther, E.L, Cao, Q, Lu, J, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2018-02-02 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4AO6
 
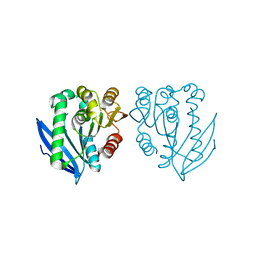 | | Native structure of a novel cold-adapted esterase from an Arctic intertidal metagenomic library | | Descriptor: | ESTERASE | | Authors: | Fu, J, Leiros, H.-K.S, Pascale, D.d, Johnson, K.A, Blencke, H.M, Landfald, B. | | Deposit date: | 2012-03-23 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Functional and Structural Studies of a Novel Cold-Adapted Esterase from an Arctic Intertidal Metagenomic Library.
Appl.Microbiol.Biotechnol., 97, 2013
|
|
6CBD
 
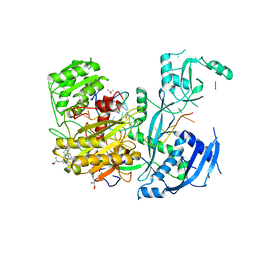 | |
4AN2
 
 | | Crystal structures of human MEK1 with carboxamide-based allosteric inhibitor XL518 (GDC-0973), or related analogs. | | Descriptor: | DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Rice, K.D, Aay, N, Anand, N.K, Blazey, C.M, Bowles, O.J, Bussenius, J, Costanzo, S, Curtis, J.K, Defina, S.C, Dubenko, L, Engst, S, Joshi, A.A, Kennedy, A.R, Kim, A.I, Koltun, E.S, Lougheed, J.C, Manalo, J.C.L, Martini, J.F, Nuss, J.M, Peto, C.J, Tsang, T.H, Yu, P, Johnston, S. | | Deposit date: | 2012-03-14 | | Release date: | 2012-12-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel Carboxamide-Based Allosteric Mek Inhibitors: Discovery and Optimization Efforts Toward Xl518 (Gdc-0973)
Acs Med.Chem.Lett., 3, 2012
|
|
5TLZ
 
 | | Fructose-1,6-bisphosphate aldolase from rabbit muscle in complex with the inhibitor naphthalene 2,6-bisphosphate | | Descriptor: | Fructose-bisphosphate aldolase A, GLYCEROL, naphthalene-2,6-diyl bis[dihydrogen (phosphate)] | | Authors: | Heron, P.W, Sygusch, J. | | Deposit date: | 2016-10-12 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Bisphosphonate Inhibitors of Mammalian Glycolytic Aldolase.
J.Med.Chem., 61, 2018
|
|
