1PDZ
 
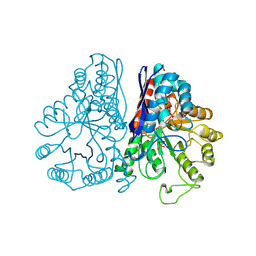 | | X-RAY STRUCTURE AND CATALYTIC MECHANISM OF LOBSTER ENOLASE | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, ENOLASE, MANGANESE (II) ION | | Authors: | Janin, J, Duquerroy, S, Camus, C, Le Bras, G. | | Deposit date: | 1995-06-05 | | Release date: | 1995-11-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structure and catalytic mechanism of lobster enolase.
Biochemistry, 34, 1995
|
|
3KGA
 
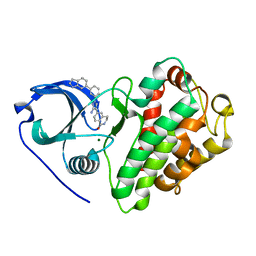 | | Crystal structure of MAPKAP kinase 2 (MK2) complexed with a potent 3-aminopyrazole ATP site inhibitor | | Descriptor: | 6-{3-amino-1-[3-(1H-indol-6-yl)phenyl]-1H-pyrazol-4-yl}-3,4-dihydroisoquinolin-1(2H)-one, MAGNESIUM ION, MAP kinase-activated protein kinase 2 | | Authors: | Kroemer, M, Velcicky, J, Izaac, A, Be, C, Huppertz, C, Pflieger, D, Schlapbach, A, Scheufler, C. | | Deposit date: | 2009-10-28 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Novel 3-aminopyrazole inhibitors of MK-2 discovered by scaffold hopping strategy.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KDM
 
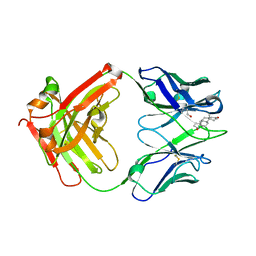 | |
4A63
 
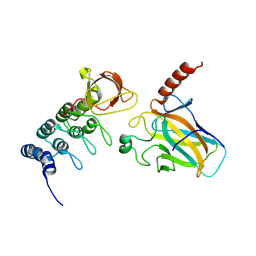 | | Crystal structure of the p73-ASPP2 complex at 2.6A resolution | | Descriptor: | ACETATE ION, APOPTOSIS STIMULATING OF P53 PROTEIN 2, TUMOUR PROTEIN 73, ... | | Authors: | Canning, P, Sharpe, T, Krojer, T, Savitsky, P, Cooper, C.D.O, Salah, E, Keates, T, Muniz, J, Vollmar, M, von Delft, F, Weigelt, J, Arrowsmith, C, Bountra, C, Edwards, A, Bullock, A.N. | | Deposit date: | 2011-10-31 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural Basis for Aspp2 Recognition by the Tumor Suppressor P73.
J.Mol.Biol., 423, 2012
|
|
3WBK
 
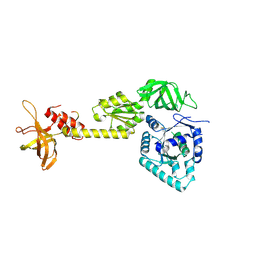 | | crystal structure analysis of eukaryotic translation initiation factor 5B and 1A complex | | Descriptor: | Eukaryotic translation initiation factor 1A, Eukaryotic translation initiation factor 5B | | Authors: | Zheng, A, Yamamoto, R, Ose, T, Yu, J, Tanaka, I, Yao, M. | | Deposit date: | 2013-05-20 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | X-ray structures of eIF5B and the eIF5B-eIF1A complex: the conformational flexibility of eIF5B is restricted on the ribosome by interaction with eIF1A
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3KF3
 
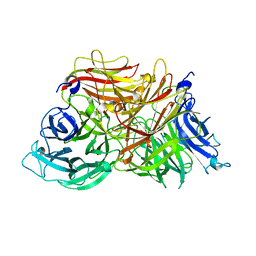 | |
3WNF
 
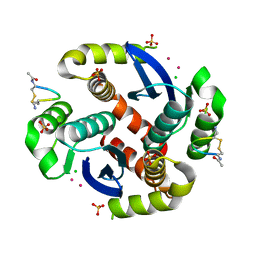 | |
3GW4
 
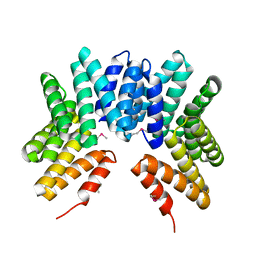 | | Crystal structure of uncharacterized protein from Deinococcus radiodurans. Northeast Structural Genomics Consortium Target DrR162B. | | Descriptor: | Uncharacterized protein | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Sahdev, S, Xiao, R, Foote, E.L, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-03-31 | | Release date: | 2009-04-21 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Northeast Structural Genomics Consortium Target DrR162B
To be published
|
|
1SSD
 
 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | SULFATE ION, Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-24 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
3WQT
 
 | | Staphylococcus aureus FtsA complexed with AMPPNP | | Descriptor: | CHLORIDE ION, Cell division protein FtsA, MAGNESIUM ION, ... | | Authors: | Fujita, J, Maeda, Y, Miyazaki, Y, Inoue, T, Matsumura, H. | | Deposit date: | 2014-02-01 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of FtsA from Staphylococcus aureus
FEBS Lett., 588, 2014
|
|
3GZD
 
 | | Human selenocysteine lyase, P1 crystal form | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, NITRATE ION, Selenocysteine lyase | | Authors: | Karlberg, T, Hogbom, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Schutz, P, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-07 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical discrimination between selenium and sulfur 1: a single residue provides selenium specificity to human selenocysteine lyase.
Plos One, 7, 2012
|
|
3W3V
 
 | |
3H8T
 
 | | Structure of Porphyromonas gingivalis heme-binding protein HmuY in complex with Heme | | Descriptor: | GLYCEROL, HmuY, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Wojtowicz, H, Guevara, T, Tallant, C, Olczak, M, Sroka, A, Potempa, J, Sola, M, Olczak, T, Gomis-Ruth, F.X. | | Deposit date: | 2009-04-29 | | Release date: | 2009-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unique structure and stability of HmuY, a novel heme-binding protein of Porphyromonas gingivalis
Plos Pathog., 5, 2009
|
|
3HA3
 
 | |
3W6G
 
 | | Structure of peroxiredoxin from anaerobic hyperthermophilic archaeon Pyrococcus horikoshii | | Descriptor: | CITRATE ANION, Probable peroxiredoxin | | Authors: | Nakamura, T, Mori, A, Niiyama, M, Matsumura, H, Tokuyama, C, Morita, J, Uegaki, K, Inoue, T. | | Deposit date: | 2013-02-14 | | Release date: | 2013-07-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of peroxiredoxin from the anaerobic hyperthermophilic archaeon Pyrococcus horikoshii
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3H6N
 
 | | Crystal Structure of the ubiquitin-like domain of plexin D1 | | Descriptor: | ARSENIC, Plexin-D1, UNKNOWN ATOM OR ION | | Authors: | Tong, Y, Nedyalkova, L, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Buck, M, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-23 | | Release date: | 2009-05-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Crystal Structure of the ubiquitin-like domain of plexin D1
To be Published
|
|
3WAS
 
 | | Crystal structure of 4-O-beta-D-mannosyl-D-glucose phosphorylase MGP complexed with Man-Glc+PO4 | | Descriptor: | 4-O-beta-D-mannosyl-D-glucose phosphorylase, PHOSPHATE ION, beta-D-mannopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Nakae, S, Ito, S, Higa, M, Senoura, T, Wasaki, J, Hijikata, A, Shionyu, M, Ito, S, Shirai, T. | | Deposit date: | 2013-05-08 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Novel Enzyme in Mannan Biodegradation Process 4-O-beta-d-Mannosyl-d-Glucose Phosphorylase MGP
J.Mol.Biol., 425, 2013
|
|
3H96
 
 | | Msmeg_3358 F420 Reductase | | Descriptor: | 1,2-ETHANEDIOL, F420-H2 Dependent Reductase A | | Authors: | Jackson, C.J, French, N, Newman, J, Taylor, M.C, Russell, R.J, Oakeshott, J.G. | | Deposit date: | 2009-04-30 | | Release date: | 2010-04-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and characterization of two families of F420 H2-dependent reductases from Mycobacteria that catalyse aflatoxin degradation.
Mol.Microbiol., 78, 2010
|
|
3HA7
 
 | |
3GJQ
 
 | | Caspase-3 Binds Diverse P4 Residues in Peptides | | Descriptor: | Caspase-3 subunit p12, Caspase-3 subunit p17, peptide inhibitor | | Authors: | Fang, B, Fu, G, Agniswamy, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2009-03-09 | | Release date: | 2009-03-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Caspase-3 binds diverse P4 residues in peptides as revealed by crystallography and structural modeling.
Apoptosis, 14, 2009
|
|
3WCY
 
 | |
4NW2
 
 | | Tandem chromodomains of human CHD1 in complex with Influenza virus NS1 C-terminal tail trimethylated at K229 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1, GLYCEROL, Nonstructural protein 1, ... | | Authors: | Qin, S, Tempel, W, Xu, C, El Bakkouri, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-05 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for histone mimicry and hijacking of host proteins by influenza virus protein NS1.
Nat Commun, 5, 2014
|
|
3H4R
 
 | | Crystal structure of E. coli RecE exonuclease | | Descriptor: | Exodeoxyribonuclease 8 | | Authors: | Bell, C.E, Zhang, J. | | Deposit date: | 2009-04-20 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of E. coli RecE protein reveals a toroidal tetramer for processing double-stranded DNA breaks.
Structure, 17, 2009
|
|
3WFT
 
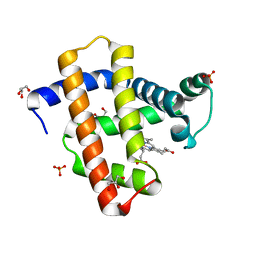 | | Crystal structure of horse heart myoglobin reconstituted with cobalt(II) tetradehydrocorrin | | Descriptor: | (1R,19R) cobalt tetradehydrocorrin, (1S,19S) cobalt tetradehydrocorrin, GLYCEROL, ... | | Authors: | Mizohata, E, Morita, Y, Oohora, K, Hirata, K, Ohbayashi, J, Inoue, T, Hisaeda, Y, Hayashi, T. | | Deposit date: | 2013-07-23 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Co(II)/Co(I) reduction-induced axial histidine-flipping in myoglobin reconstituted with a cobalt tetradehydrocorrin as a methionine synthase model.
Chem.Commun.(Camb.), 50, 2014
|
|
3WG1
 
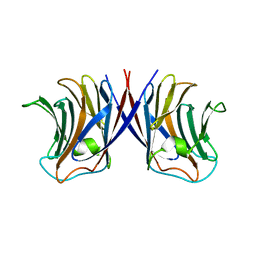 | | Crystal structure of Agrocybe cylindracea galectin with lactose | | Descriptor: | Galactoside-binding lectin, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Kuwabara, N, Hu, D, Tateno, H, Makio, H, Hirabayashi, J, Kato, R. | | Deposit date: | 2013-07-25 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational change of a unique sequence in a fungal galectin from Agrocybe cylindracea controls glycan ligand-binding specificity.
Febs Lett., 587, 2013
|
|
