3VKF
 
 | | Crystal Structure of Neurexin 1beta/Neuroligin 1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neurexin-1-beta, ... | | Authors: | Tanaka, H, Miyazaki, N, Nogi, T, Iwasaki, K, Takagi, J. | | Deposit date: | 2011-11-15 | | Release date: | 2012-08-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Higher-order architecture of cell adhesion mediated by polymorphic synaptic adhesion molecules neurexin and neuroligin.
Cell Rep, 2, 2012
|
|
3HH7
 
 | | Structural and Functional Characterization of a Novel Homodimeric Three-finger Neurotoxin from the Venom of Ophiophagus hannah (King Cobra) | | Descriptor: | Muscarinic toxin-like protein 3 homolog | | Authors: | Roy, A, Zhou, X, Chong, M.Z, D'hoedt, D, Foo, C.S, Rajagopalan, N, Nirthanan, S, Bertrand, D, Sivaraman, J, Kini, R.M. | | Deposit date: | 2009-05-15 | | Release date: | 2010-01-12 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and Functional Characterization of a Novel Homodimeric Three-finger Neurotoxin from the Venom of Ophiophagus hannah (King Cobra)
J.Biol.Chem., 285, 2010
|
|
1QCR
 
 | | CRYSTAL STRUCTURE OF BOVINE MITOCHONDRIAL CYTOCHROME BC1 COMPLEX, ALPHA CARBON ATOMS ONLY | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, UBIQUINOL CYTOCHROME C OXIDOREDUCTASE | | Authors: | Xia, D, Yu, C.A, Kim, H, Xia, J.Z, Kachurin, A, Zhang, L, Yu, L, Deisenhofer, J. | | Deposit date: | 1997-05-17 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria.
Science, 277, 1997
|
|
1QMY
 
 | | FMDV LEADER PROTEASE (LBSHORT-C51A-C133S) | | Descriptor: | 1,2-ETHANEDIOL, PROTEASE | | Authors: | Guarne, A, Tormo, J, Glaser, W, Skern, T, Fita, I. | | Deposit date: | 1999-10-08 | | Release date: | 2000-10-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Biochemical Features Distinguish the Foot-and-Mouth Disease Virus Leader Proteinase from Other Papain-Like Enzymes
J.Mol.Biol., 302, 2000
|
|
3VPA
 
 | | Staphylococcus aureus FtsZ apo-form | | Descriptor: | Cell division protein FtsZ | | Authors: | Matsui, T, Yamane, J, Mogi, N, Yao, M, Tanaka, I. | | Deposit date: | 2012-02-28 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Structural reorganization of the bacterial cell-division protein FtsZ from Staphylococcus aureus
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3HFW
 
 | | Crystal Structure of human ADP-ribosylhydrolase 1 (hARH1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Mueller-Dieckmann, C, Weiss, M.S, Mueller-Dieckmann, J, Koch-Nolte, F. | | Deposit date: | 2009-05-13 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of human ADP-ribosylhydrolase 1
To be Published
|
|
3VPV
 
 | |
3HHD
 
 | | Structure of the Human Fatty Acid Synthase KS-MAT Didomain as a Framework for Inhibitor Design. | | Descriptor: | CHLORIDE ION, Fatty acid synthase | | Authors: | Pappenberger, G.M, Benz, J, Thoma, R, Rudolph, M.G. | | Deposit date: | 2009-05-15 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the human fatty acid synthase KS-MAT didomain as a framework for inhibitor design.
J.Mol.Biol., 397, 2010
|
|
3HIL
 
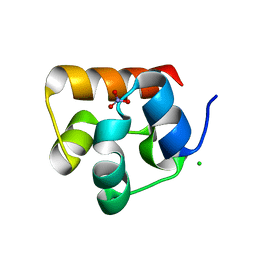 | | SAM Domain of Human Ephrin Type-A Receptor 1 (EphA1) | | Descriptor: | CHLORIDE ION, Ephrin type-A receptor 1, NITRATE ION | | Authors: | Walker, J.R, Yermekbayeva, L, Butler-Cole, C, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-05-20 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SAM Domain of Human Ephrin Type-A Receptor 1 (EphA1).
To be Published
|
|
4LCC
 
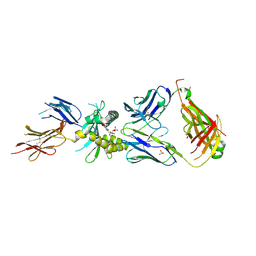 | | Crystal structure of a human MAIT TCR in complex with a bacterial antigen bound to humanized bovine MR1 | | Descriptor: | 1-deoxy-1-[6-(hydroxymethyl)-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl]-D-arabinitol, Beta-2-microglobulin, MHC class I-related protein, ... | | Authors: | Lopez-Sagaseta, J, Adams, E.J. | | Deposit date: | 2013-06-21 | | Release date: | 2013-10-16 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3.263 Å) | | Cite: | MAIT Recognition of a Stimulatory Bacterial Antigen Bound to MR1.
J.Immunol., 191, 2013
|
|
3VBC
 
 | |
3VWK
 
 | |
3VBV
 
 | |
3HL3
 
 | | 2.76 Angstrom Crystal Structure of a Putative Glucose-1-Phosphate Thymidylyltransferase from Bacillus anthracis in Complex with a Sucrose. | | Descriptor: | Glucose-1-phosphate thymidylyltransferase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Minasov, G, Shuvalova, L, Halavaty, A, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-26 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | 2.76 Angstrom Crystal Structure of a Putative Glucose-1-Phosphate Thymidylyltransferase from Bacillus anthracis in Complex with a Sucrose.
TO BE PUBLISHED
|
|
3VXG
 
 | | Crystal structure of conjugated polyketone reductase C2 from Candida Parapsilosis | | Descriptor: | Conjugated polyketone reductase C2 | | Authors: | Qin, H.-M, Yamamura, A, Miyakawa, T, Maruoka, S, Ohtsuka, J, Nagata, K, Kataoka, M, Shimizu, S, Tanokura, M. | | Deposit date: | 2012-09-13 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of conjugated polyketone reductase from Candida parapsilosis IFO 0708 reveals conformational changes for substrate recognition upon NADPH binding
Appl.Microbiol.Biotechnol., 98, 2014
|
|
3HL4
 
 | | Crystal structure of a mammalian CTP:phosphocholine cytidylyltransferase with CDP-choline | | Descriptor: | Choline-phosphate cytidylyltransferase A, FORMIC ACID, GLYCEROL, ... | | Authors: | Lee, J, Paetzel, M, Cornell, R.B. | | Deposit date: | 2009-05-26 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a mammalian CTP: Phosphocholine cytidylyltransferase catalytic domain reveals novel active site residues within a highly conserved nucleotidyl-transferase fold
J.Biol.Chem., 284, 2009
|
|
3VEQ
 
 | | A binary complex betwwen bovine pancreatic trypsin and a engineered mutant trypsin inhibitor | | Descriptor: | CALCIUM ION, Cationic trypsin, Chymotrypsin inhibitor 3 | | Authors: | Sen, U, Majumder, S, Khamrui, S, Dasgupta, J. | | Deposit date: | 2012-01-09 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Role of remote scaffolding residues in the inhibitory loop pre-organization, flexibility, rigidification and enzyme inhibition of serine protease inhibitors
Biochim.Biophys.Acta, 1824, 2012
|
|
3HL6
 
 | | Staphylococcus aureus pathogenicity island 3 ORF9 protein | | Descriptor: | CHLORIDE ION, Pathogenicity island protein | | Authors: | Kruse, A.C, Huseby, M, Shi, K, Digre, J, Schlievert, P.M, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2009-05-26 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional studies of a pathogenicity island protein from Staphylococcus aureus
To be Published
|
|
3VIC
 
 | | Green-fluorescent variant of the non-fluorescent chromoprotein Rtms5 | | Descriptor: | CHLORIDE ION, GFP-like non-fluorescent chromoprotein, IODIDE ION | | Authors: | Battad, J.M, Traore, D.A.K, Byres, E, Wilce, M, Devenish, R.J, Rossjohn, J, Prescott, M. | | Deposit date: | 2011-09-28 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Green Fluorescent Protein Containing a QFG Tri-Peptide Chromophore: Optical Properties and X-Ray Crystal Structure.
Plos One, 7, 2012
|
|
3HG0
 
 | | Crystal structure of a DARPin in complex with ORF49 from Lactococcal phage TP901-1 | | Descriptor: | Baseplate protein, Designed Ankyrin Repeat Protein (DARPin) 20 | | Authors: | Veesler, D, Dreier, B, Blangy, S, Lichiere, J, Tremblay, D, Moineau, S, Spinelli, S, Tegoni, M, Pluckthun, A, Campanacci, V, Cambillau, C. | | Deposit date: | 2009-05-13 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and function of a DARPin neutralizing inhibitor of lactococcal phage TP901-1: comparison of DARPin and camelid VHH binding mode.
J.Biol.Chem., 284, 2009
|
|
3VLN
 
 | | Human Glutathione Transferase O1-1 C32S Mutant in Complex with Ascorbic Acid | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ASCORBIC ACID, ... | | Authors: | Brock, J, Board, P.G, Oakley, A.J. | | Deposit date: | 2011-12-02 | | Release date: | 2012-05-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the dehydroascorbate reductase activity of human omega-class glutathione transferases.
J.Mol.Biol., 420, 2012
|
|
3HL9
 
 | | Simvastatin Synthase (LovD) from Aspergillus terreus, unliganded | | Descriptor: | Transesterase | | Authors: | Sawaya, M.R, Yeates, T.O, Laidman, J, Pashkov, I, Gao, X, Tang, Y. | | Deposit date: | 2009-05-27 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Directed evolution and structural characterization of a simvastatin synthase
Chem.Biol., 16, 2009
|
|
3VBQ
 
 | |
3HHP
 
 | |
3VOZ
 
 | | Crystal structure of human glutaminase in complex with BPTES | | Descriptor: | Glutaminase kidney isoform, mitochondrial, N,N'-[sulfanediylbis(ethane-2,1-diyl-1,3,4-thiadiazole-5,2-diyl)]bis(2-phenylacetamide), ... | | Authors: | Thangavelu, K, Sivaraman, J. | | Deposit date: | 2012-02-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
