8E0W
 
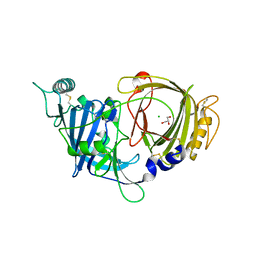 | | Crystal structure of mouse APCDD1 in P1 space group | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Hsieh, F.L, Chang, T.H, Gabelli, S.B, Nathans, J. | | Deposit date: | 2022-08-09 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of WNT inhibitor adenomatosis polyposis coli down-regulated 1 (APCDD1), a cell-surface lipid-binding protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8DST
 
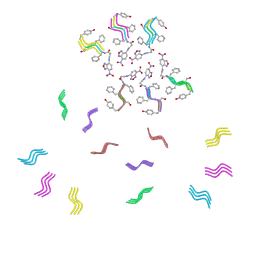 | |
8E0R
 
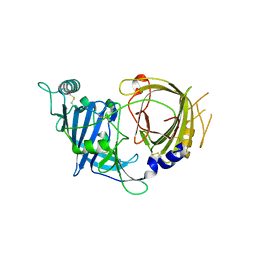 | | Crystal structure of mouse APCDD1 in P21 space group | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein APCDD1 | | Authors: | Hsieh, F.L, Chang, T.H, Gabelli, S.B, Nathans, J. | | Deposit date: | 2022-08-09 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of WNT inhibitor adenomatosis polyposis coli down-regulated 1 (APCDD1), a cell-surface lipid-binding protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8DL5
 
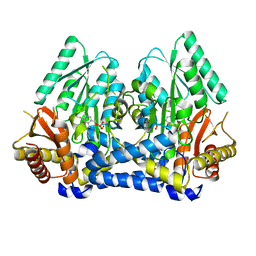 | |
4ABH
 
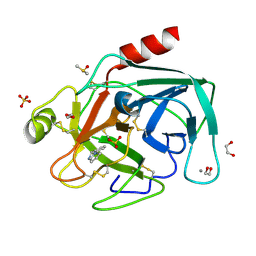 | | Fragments bound to bovine trypsin for the SAMPL challenge | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-PYRROLIDIN-1-YLPHENYL)METHANAMINE, CALCIUM ION, ... | | Authors: | Newman, J, Peat, T.S. | | Deposit date: | 2011-12-08 | | Release date: | 2012-02-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The Dingo Dataset: A Comprehensive Set of Data for the Sampl Challenge.
J.Comput.Aided Mol.Des., 26, 2012
|
|
6NPR
 
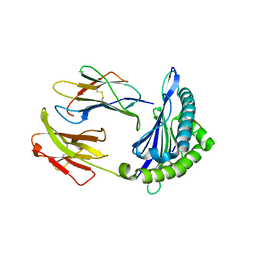 | | Crystal structure of H-2Dd with C84-C139 disulfide in complex with gp120 derived peptide P18-I10 | | Descriptor: | ARG-GLY-PRO-GLY-ARG-ALA-PHE-VAL-THR-ILE, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Toor, J, McShan, A.C, Tripathi, S.M, Sgourakis, N.G. | | Deposit date: | 2019-01-18 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Molecular determinants of chaperone interactions on MHC-I for folding and antigen repertoire selection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4I07
 
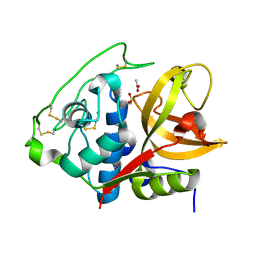 | | Structure of mature form of cathepsin B1 from Schistosoma mansoni | | Descriptor: | ACETATE ION, CHLORIDE ION, Cathepsin B-like peptidase (C01 family) | | Authors: | Rezacova, P, Jilkova, A, Brynda, J, Horn, M, Mares, M. | | Deposit date: | 2012-11-16 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Activation route of the Schistosoma mansoni cathepsin B1 drug target: structural map with a glycosaminoglycan switch
Structure, 22, 2014
|
|
421P
 
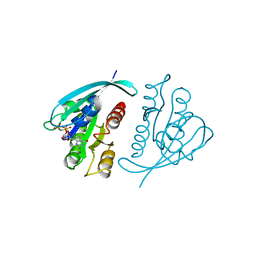 | | THREE-DIMENSIONAL STRUCTURES OF H-RAS P21 MUTANTS: MOLECULAR BASIS FOR THEIR INABILITY TO FUNCTION AS SIGNAL SWITCH MOLECULES | | Descriptor: | H-RAS P21 PROTEIN, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Krengel, U, John, J, Scherer, A, Kabsch, W, Wittinghofer, A, Pai, E.F. | | Deposit date: | 1991-06-06 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structures of H-ras p21 mutants: molecular basis for their inability to function as signal switch molecules.
Cell(Cambridge,Mass.), 62, 1990
|
|
8E4T
 
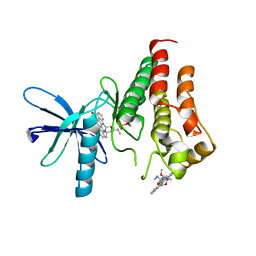 | | Crystal structure of the kinase domain of RTKC8 from the choanoflagellate Monosiga brevicollis | | Descriptor: | PHOSPHATE ION, RTKC8 Kinase domain, STAUROSPORINE | | Authors: | Bajaj, T, Gee, C.L, Kuriyan, J. | | Deposit date: | 2022-08-18 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the kinase domain of a receptor tyrosine kinase from a choanoflagellate, Monosiga brevicollis.
Plos One, 18, 2023
|
|
1MWK
 
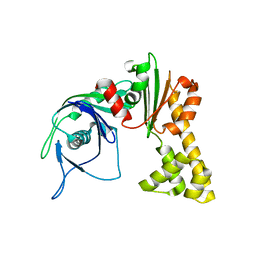 | | ParM from plasmid R1 APO form | | Descriptor: | ParM | | Authors: | Van den Ent, F, Moller-Jensen, J, Amos, L.A, Gerdes, K, Lowe, J. | | Deposit date: | 2002-09-30 | | Release date: | 2003-01-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | F-actin-like filaments formed by plasmid segregation protein ParM
EMBO J., 21, 2002
|
|
8DU2
 
 | | HnRNPA2 D290V LCD PM1 | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1 | | Authors: | Eisenberg, D.S, Lu, J, Ge, P, Boyer, D.R. | | Deposit date: | 2022-07-26 | | Release date: | 2023-08-02 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the D290V mutant of the hnRNPA2 low-complexity domain suggests how D290V affects phase separation and aggregation.
J.Biol.Chem., 300, 2023
|
|
8DSV
 
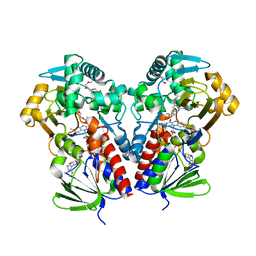 | | The structure of NicA2 in complex with N-methylmyosmine | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FAD-dependent oxidoreductase, ... | | Authors: | Wu, K, Dulchavsky, M, Li, J, Bardwell, J.C.A. | | Deposit date: | 2022-07-22 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Directed evolution unlocks oxygen reactivity for a nicotine-degrading flavoenzyme.
Nat.Chem.Biol., 19, 2023
|
|
8DQ8
 
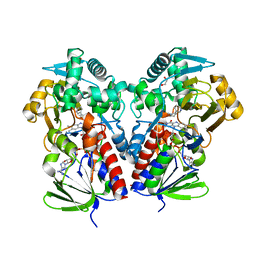 | | The structure of NicA2 variant F104L/A107T/S146I/G317D/H368R/L449V/N462S in complex with N-methylmyosmine | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, 1,2-ETHANEDIOL, Amine oxidase, ... | | Authors: | Wu, K, Dulchavsky, M, Li, J, Bardwell, J.C.A. | | Deposit date: | 2022-07-18 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed evolution unlocks oxygen reactivity for a nicotine-degrading flavoenzyme.
Nat.Chem.Biol., 19, 2023
|
|
8DQ7
 
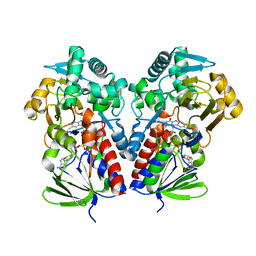 | |
8DSP
 
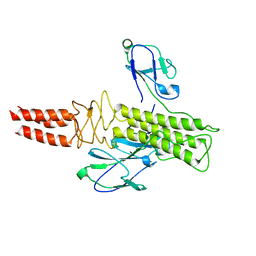 | |
8DUW
 
 | | HnRNPA2 D290V LCD PM2 | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1 | | Authors: | Eisenberg, D.S, Lu, J, Ge, P, Boyer, D.R. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-02 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of the D290V mutant of the hnRNPA2 low-complexity domain suggests how D290V affects phase separation and aggregation.
J.Biol.Chem., 300, 2023
|
|
2NV4
 
 | | Crystal structure of UPF0066 protein AF0241 in complex with S-adenosylmethionine. Northeast Structural Genomics Consortium target GR27 | | Descriptor: | ACETATE ION, S-ADENOSYLMETHIONINE, UPF0066 protein AF_0241 | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Vorobiev, S.M, Fang, Y, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-10 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional insights from structural genomics.
J.Struct.Funct.Genom., 8, 2007
|
|
6LUF
 
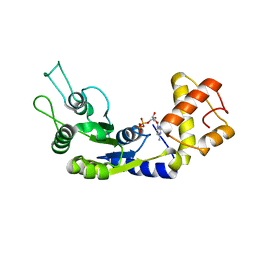 | |
8DLA
 
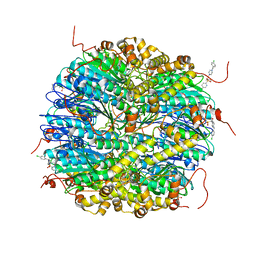 | | ClpP2 from Chlamydia trachomatis bound by MAS1-12 | | Descriptor: | 1-{4-[(4-chlorophenyl)methyl]piperazin-1-yl}-2-methyl-2-[5-(trifluoromethyl)pyridine-2-sulfonyl]propan-1-one, ATP-dependent Clp protease proteolytic subunit 2, GLYCEROL, ... | | Authors: | Azadmanesh, J, Struble, L.R, Seleem, M.A, Ouellette, S, Conda-Sheridan, M, Borgstahl, G.E.O. | | Deposit date: | 2022-07-07 | | Release date: | 2023-08-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | The structure of caseinolytic protease subunit ClpP2 reveals a functional model of the caseinolytic protease system from Chlamydia trachomatis.
J.Biol.Chem., 299, 2023
|
|
1XGB
 
 | |
2OJW
 
 | | Crystal structure of human glutamine synthetase in complex with ADP and phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Karlberg, T, Uppenberg, J, Arrowsmith, C, Berglund, H, Busam, R.D, Collins, R, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Hogbom, M, Johansson, I, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Wallden, K, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-15 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of mammalian glutamine synthetases illustrate substrate-induced conformational changes and provide opportunities for drug and herbicide design.
J.Mol.Biol., 375, 2008
|
|
1XGC
 
 | |
1L8O
 
 | | Molecular basis for the local conformational rearrangement of human phosphoserine phosphatase | | Descriptor: | L-3-phosphoserine phosphatase, PHOSPHATE ION, SERINE | | Authors: | Kim, H.Y, Heo, Y.S, Kim, J.H, Park, M.H, Moon, J, Park, S.Y, Lee, T.G, Jeon, Y.H, Ro, S, Hwang, K.Y. | | Deposit date: | 2002-03-21 | | Release date: | 2003-04-01 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for the local conformational rearrangement of human phosphoserine phosphatase
J.Biol.Chem., 277, 2002
|
|
2O2T
 
 | | The crystal structure of the 1st PDZ domain of MPDZ | | Descriptor: | Multiple PDZ domain protein | | Authors: | Papagrigoriou, E, Gileadi, C, Phillips, C, Johansson, C, Salah, E, Savitsky, P, Gorrec, F, Umeano, C, Berridge, G, Pike, A.C.W, Elkins, J, Edwards, A, Arrowsmith, C, Weigelt, J, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-30 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of the 1st PDZ domain of MPDZ
To be Published
|
|
8EC7
 
 | | HnRNPA2 D290V LCD PM3 | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1 | | Authors: | Eisenberg, D.S, Lu, J, Ge, P, Boyer, D.R. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of the D290V mutant of the hnRNPA2 low-complexity domain suggests how D290V affects phase separation and aggregation.
J.Biol.Chem., 300, 2023
|
|
