3HFG
 
 | |
4FPW
 
 | | Crystal Structure of CalU16 from Micromonospora echinospora. Northeast Structural Genomics Consortium Target MiR12. | | Descriptor: | CalU16 | | Authors: | Seetharaman, J, Lew, S, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Phillips Jr, G.N, Kennedy, M.A, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-22 | | Release date: | 2012-12-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Guided Functional Characterization of Enediyne Self-Sacrifice Resistance Proteins, CalU16 and CalU19.
Acs Chem.Biol., 9, 2014
|
|
1BU4
 
 | | RIBONUCLEASE 1 COMPLEX WITH 2'GMP | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1 | | Authors: | Loris, R, Devos, S, Langhorst, U, Decanniere, K, Bouckaert, J, Maes, D, Transue, T.R, Steyaert, J. | | Deposit date: | 1998-09-11 | | Release date: | 1999-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conserved water molecules in a large family of microbial ribonucleases.
Proteins, 36, 1999
|
|
3HJD
 
 | |
1BKB
 
 | | INITIATION FACTOR 5A FROM ARCHEBACTERIUM PYROBACULUM AEROPHILUM | | Descriptor: | TRANSLATION INITIATION FACTOR 5A | | Authors: | Peat, T.S, Newman, J, Waldo, G.S, Berendzen, J, Terwilliger, T.C. | | Deposit date: | 1998-07-05 | | Release date: | 1998-11-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of translation initiation factor 5A from Pyrobaculum aerophilum at 1.75 A resolution.
Structure, 6, 1998
|
|
3HJ4
 
 | | Minor Editosome-Associated TUTase 1 | | Descriptor: | Minor Editosome-Associated TUTase | | Authors: | Stagno, J, Luecke, H. | | Deposit date: | 2009-05-20 | | Release date: | 2010-05-12 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of the Mitochondrial Editosome-Like Complex Associated TUTase 1 Reveals Divergent Mechanisms of UTP Selection and Domain Organization.
J.Mol.Biol., 399, 2010
|
|
3CFI
 
 | | Nanobody-aided structure determination of the EPSI:EPSJ pseudopilin heterdimer from Vibrio Vulnificus | | Descriptor: | CHLORIDE ION, Nanobody NBEPSIJ_11, Type II secretory pathway, ... | | Authors: | Lam, A.Y, Pardon, E, Korotkov, K.V, Steyaert, J, Hol, W.G.J. | | Deposit date: | 2008-03-03 | | Release date: | 2009-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Nanobody-aided structure determination of the EpsI:EpsJ pseudopilin heterodimer from Vibrio vulnificus.
J.Struct.Biol., 166, 2009
|
|
3CGN
 
 | | Crystal Structure of thermophilic SlyD | | Descriptor: | Peptidyl-prolyl cis-trans isomerase, SULFATE ION | | Authors: | Neumann, P, Loew, C, Stubbs, M.T, Balbach, J. | | Deposit date: | 2008-03-06 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure Determination and Functional Characterization of the Metallochaperone SlyD from Thermus thermophilus
J.Mol.Biol., 398, 2010
|
|
1BPH
 
 | | CONFORMATIONAL CHANGES IN CUBIC INSULIN CRYSTALS IN THE PH RANGE 7-11 | | Descriptor: | 1,2-DICHLOROETHANE, INSULIN A CHAIN (PH 9), INSULIN B CHAIN (PH 9), ... | | Authors: | Gursky, O, Badger, J, Li, Y, Caspar, D.L.D. | | Deposit date: | 1992-10-30 | | Release date: | 1993-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational changes in cubic insulin crystals in the pH range 7-11.
Biophys.J., 63, 1992
|
|
1BR0
 
 | | THREE DIMENSIONAL STRUCTURE OF THE N-TERMINAL DOMAIN OF SYNTAXIN 1A | | Descriptor: | PROTEIN (SYNTAXIN 1-A) | | Authors: | Fernandez, I, Ubach, J, Dubulova, I, Zhang, X, Sudhof, T.C, Rizo, J. | | Deposit date: | 1998-08-25 | | Release date: | 1998-09-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of an evolutionarily conserved N-terminal domain of syntaxin 1A.
Cell(Cambridge,Mass.), 94, 1998
|
|
2QSD
 
 | | Crystal structure of a protein Il1583 from Idiomarina loihiensis | | Descriptor: | GLYCEROL, Uncharacterized conserved protein | | Authors: | Patskovsky, Y, Bonanno, J, Sauder, J.M, Romero, R, Rutter, M, Koss, J, Mckenzie, C, Gheyi, T, Bain, K, Wasserman, S.R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-07-30 | | Release date: | 2007-08-14 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Protein Il1583 from Idiomarina loihiensis.
To be Published
|
|
3VQ5
 
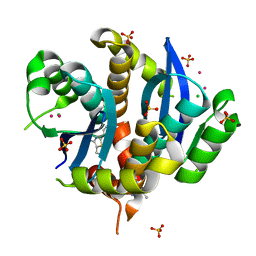 | | HIV-1 IN core domain in complex with N-METHYL-1-(4-METHYL-2-PHENYL-1,3-THIAZOL-5-YL)METHANAMINE | | Descriptor: | CADMIUM ION, CHLORIDE ION, N-methyl-1-(4-methyl-2-phenyl-1,3-thiazol-5-yl)methanamine, ... | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-20 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
2HED
 
 | | CONTRIBUTION OF WATER MOLECULES IN THE INTERIOR OF A PROTEIN TO THE CONFORMATIONAL STABILITY | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Funahashi, J, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1997-09-16 | | Release date: | 1998-01-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of water molecules in the interior of a protein to the conformational stability.
J.Mol.Biol., 274, 1997
|
|
4AU0
 
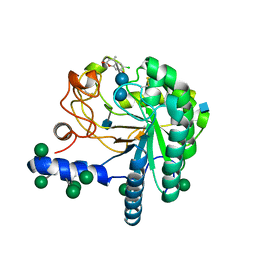 | | Hypocrea jecorina Cel6A D221A mutant soaked with 6-chloro-4- methylumbelliferyl-beta-cellobioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-chloro-7-hydroxy-4-methyl-2H-chromen-2-one, EXOGLUCANASE 2, ... | | Authors: | Wu, M, Nerinckx, W, Piens, K, Ishida, T, Hansson, H, Stahlberg, J, Sandgren, M. | | Deposit date: | 2012-05-11 | | Release date: | 2013-01-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Design, Synthesis, Evaluation and Enzyme-Substrate Structures of Improved Fluorogenic Substrates for Family 6 Glycoside Hydrolases.
FEBS J., 280, 2013
|
|
1C15
 
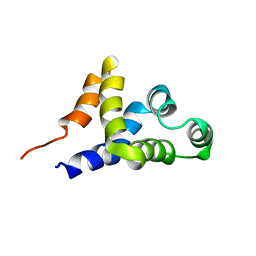 | | SOLUTION STRUCTURE OF APAF-1 CARD | | Descriptor: | APOPTOTIC PROTEASE ACTIVATING FACTOR 1 | | Authors: | Zhou, P, Chou, J, Olea, R.S, Yuan, J, Wagner, G. | | Deposit date: | 1999-07-20 | | Release date: | 1999-09-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Apaf-1 CARD and its interaction with caspase-9 CARD: a structural basis for specific adaptor/caspase interaction.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1BNL
 
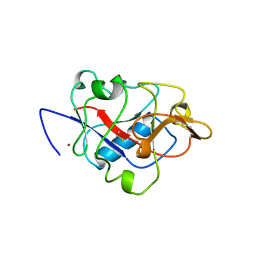 | | ZINC DEPENDENT DIMERS OBSERVED IN CRYSTALS OF HUMAN ENDOSTATIN | | Descriptor: | COLLAGEN XVIII, ZINC ION | | Authors: | Ding, Y.-H, Javaherian, K, Lo, K.-M, Chopra, R, Boehm, T, Lanciotti, J, Harris, B.A, Li, Y, Shapiro, R, Hohenester, E, Timpl, R, Folkman, J, Wiley, D.C. | | Deposit date: | 1998-07-30 | | Release date: | 1998-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Zinc-dependent dimers observed in crystals of human endostatin.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
3CU1
 
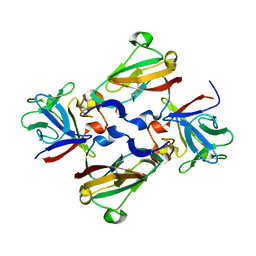 | | Crystal Structure of 2:2:2 FGFR2D2:FGF1:SOS complex | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, Fibroblast growth factor receptor 2, Heparin-binding growth factor 1 | | Authors: | Guo, F, Dakshinamurthy, R, Thallapuranam, S.K.K, Sakon, J. | | Deposit date: | 2008-04-15 | | Release date: | 2009-04-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of 2:2:2 FGFR2D2:FGF1:SOS complex
To be Published
|
|
1BPY
 
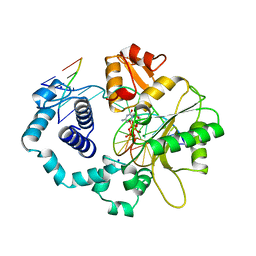 | | HUMAN DNA POLYMERASE BETA COMPLEXED WITH GAPPED DNA AND DDCTP | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*DOC)-3'), ... | | Authors: | Sawaya, M.R, Pelletier, H, Prasad, R, Wilson, S.H, Kraut, J. | | Deposit date: | 1997-04-15 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism.
Biochemistry, 36, 1997
|
|
3HMX
 
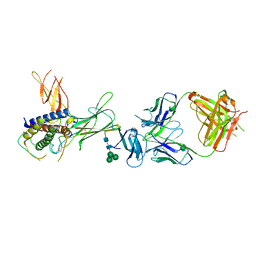 | | Crystal structure of ustekinumab FAB/IL-12 complex | | Descriptor: | Interleukin-12 subunit alpha, Interleukin-12 subunit beta, USTEKINUMAB FAB HEAVY CHAIN, ... | | Authors: | Luo, J. | | Deposit date: | 2009-05-29 | | Release date: | 2010-06-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the dual recognition of IL-12 and IL-23 by ustekinumab.
J.Mol.Biol., 402, 2010
|
|
1BWJ
 
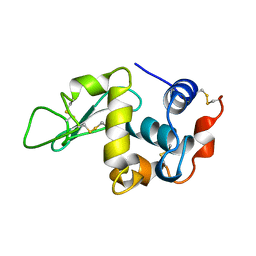 | | THE 1.8 A STRUCTURE OF MICROGRAVITY GROWN TETRAGONAL HEN EGG WHITE LYSOZYME | | Descriptor: | PROTEIN (LYSOZYME) | | Authors: | Dong, J, Boggon, T.J, Chayen, N.E, Raftery, J, Bi, R.C. | | Deposit date: | 1998-09-18 | | Release date: | 1998-09-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bound-solvent structures for microgravity-, ground control-, gel- and microbatch-grown hen egg-white lysozyme crystals at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
4I6K
 
 | | Crystal structure of probable 2-PYRONE-4,6-DICARBOXYLIC ACID HYDROLASE ABAYE1769 (TARGET EFI-505029) from Acinetobacter baumannii with citric acid bound | | Descriptor: | Amidohydrolase family protein, CITRIC ACID | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N.F, Stead, M, Love, J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-11-29 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.276 Å) | | Cite: | CRYSTAL STRUCTURE OF PROBABLE 2-PYRONE-4,6-DICARBOXYLIC ACID HYDROLASE (TARGET EFI-505029) FROM Acinetobacter baumannii
To be Published
|
|
2P0V
 
 | | Crystal structure of BT3781 protein from Bacteroides thetaiotaomicron, Northeast Structural Genomics Target BtR58 | | Descriptor: | Hypothetical protein BT3781 | | Authors: | Forouhar, F, Chen, Y, Seetharaman, J, Janjua, H, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-03-01 | | Release date: | 2007-03-20 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of BT3781 protein from Bacteroides thetaiotaomicron.
To be Published
|
|
2P6G
 
 | |
2P27
 
 | | Crystal Structure of Human Pyridoxal Phosphate Phosphatase with Mg2+ at 1.9 A resolution | | Descriptor: | MAGNESIUM ION, Pyridoxal phosphate phosphatase | | Authors: | Ramagopal, U.A, Freeman, J, Izuka, M, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-07 | | Release date: | 2007-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
2P8T
 
 | | Hypothetical protein PH0730 from Pyrococcus horikoshii OT3 | | Descriptor: | Hypothetical protein PH0730 | | Authors: | Chen, L, Zhao, M, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhu, J, Swindell, J.T, Fu, Z.-Q, Chrzas, J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-23 | | Release date: | 2007-04-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hypothetical protein PH0730 from Pyrococcus horikoshii OT3
To be Published
|
|
