6POK
 
 | | Crystal structure of the Robo3 FN2-3 domains | | Descriptor: | GLYCEROL, Roundabout homolog 3 | | Authors: | Wang, J, Pak, J.S, Ozkan, E. | | Deposit date: | 2019-07-04 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | NELL2-Robo3 complex structure reveals mechanisms of receptor activation for axon guidance.
Nat Commun, 11, 2020
|
|
6PPH
 
 | | Kaposi's sarcoma-associated herpesvirus (KSHV), C1 penton vertex register, CATC-binding structure | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Gong, D, Dai, X, Jih, J, Liu, Y.T, Bi, G.Q, Sun, R, Zhou, Z.H. | | Deposit date: | 2019-07-06 | | Release date: | 2019-09-11 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | DNA-Packing Portal and Capsid-Associated Tegument Complexes in the Tumor Herpesvirus KSHV.
Cell, 178, 2019
|
|
3UWC
 
 | | Structure of an aminotransferase (DegT-DnrJ-EryC1-StrS family) from Coxiella burnetii in complex with PMP | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Cheung, J, Franklin, M, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-12-01 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
2H8K
 
 | | Human Sulfotranferase SULT1C3 in complex with PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, SULT1C3 splice variant d | | Authors: | Tempel, W, Dombrovski, L, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-07 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and chemical profiling of the human cytosolic sulfotransferases.
Plos Biol., 5, 2007
|
|
8BMX
 
 | | Bacteroides thetaiotaomicron B12 TonB dependent transporter in complex with a surface lipoprotein | | Descriptor: | Putative TonB-linked outer membrane receptor, YncE family protein | | Authors: | Abellon-Ruiz, J, Jana, K, Silale, A, Basle, A, Kleinekathofer, U, van den Berg, B. | | Deposit date: | 2022-11-11 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.72 Å) | | Cite: | BtuB TonB-dependent transporters and BtuG surface lipoproteins form stable complexes for vitamin B 12 uptake in gut Bacteroides.
Nat Commun, 14, 2023
|
|
5UIY
 
 | | Structure of Bromodomain from human BAZ1A | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Bromodomain adjacent to zinc finger domain protein 1A | | Authors: | Oppikofer, M, Sudhamsu, J. | | Deposit date: | 2017-01-16 | | Release date: | 2017-07-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.687 Å) | | Cite: | Non-canonical reader modules of BAZ1A promote recovery from DNA damage.
Nat Commun, 8, 2017
|
|
6PEN
 
 | | Structure of Spastin Hexamer (whole model) in complex with substrate peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, EYEYEYEYEY, ... | | Authors: | Han, H, Schubert, H.L, McCullough, J, Monroe, N, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2019-06-20 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of spastin bound to a glutamate-rich peptide implies a hand-over-hand mechanism of substrate translocation.
J.Biol.Chem., 295, 2020
|
|
5UK0
 
 | | CryoEM structure of an influenza virus receptor-binding site antibody-antigen interface - Class 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Liu, Y, Pan, J, Caradonna, T, Jenni, S, Raymond, D.D, Schmidt, A.G, Harrison, S.C, Grigorieff, N. | | Deposit date: | 2017-01-19 | | Release date: | 2017-05-31 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface.
J. Mol. Biol., 429, 2017
|
|
6POG
 
 | | Crystal structure of the NELL2 EGF1-6-Robo3 FN1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, J, Pak, J.S, Ozkan, E. | | Deposit date: | 2019-07-03 | | Release date: | 2020-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.755 Å) | | Cite: | NELL2-Robo3 complex structure reveals mechanisms of receptor activation for axon guidance.
Nat Commun, 11, 2020
|
|
4A54
 
 | | Structural basis of the Dcp1:Dcp2 mRNA decapping complex activation by Edc3 and Scd6 | | Descriptor: | EDC3, MRNA DECAPPING COMPLEX SUBUNIT 2 | | Authors: | Fromm, S.A, Truffault, V, Kamenz, J, Braun, J.E, Hoffmann, N.A, Izaurralde, E, Sprangers, R. | | Deposit date: | 2011-10-24 | | Release date: | 2012-02-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Structural Basis of Edc3- and Scd6-Mediated Activation of the Dcp1:Dcp2 Mrna Decapping Complex.
Embo J., 31, 2011
|
|
4I42
 
 | | E.coli. 1,4-dihydroxy-2-naphthoyl coenzyme A synthase (ecMenB) in complex with 1-hydroxy-2-naphthoyl-CoA | | Descriptor: | 1,2-ETHANEDIOL, 1,4-Dihydroxy-2-naphthoyl-CoA synthase, 1-hydroxy-2-naphthoyl-CoA, ... | | Authors: | Sun, Y, Song, H, Li, J, Li, Y, Jiang, M, Zhou, J, Guo, Z. | | Deposit date: | 2012-11-27 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Structural basis of the induced-fit mechanism of 1,4-dihydroxy-2-naphthoyl coenzyme A synthase from the crotonase fold superfamily
Plos One, 8, 2013
|
|
3BZH
 
 | | Crystal structure of human ubiquitin-conjugating enzyme E2 E1 | | Descriptor: | GLYCEROL, Ubiquitin-conjugating enzyme E2 E1 | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-18 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
1GG9
 
 | | CRYSTAL STRUCTURE OF CATALASE HPII FROM ESCHERICHIA COLI, HIS128ASN VARIANT. | | Descriptor: | CATALASE HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Melik-Adamyan, W.R, Bravo, J, Carpena, X, Switala, J, Mate, M.J, Fita, I, Loewen, P.C. | | Deposit date: | 2000-08-11 | | Release date: | 2000-08-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Substrate flow in catalases deduced from the crystal structures of active site variants of HPII from Escherichia coli.
Proteins, 44, 2001
|
|
6PPI
 
 | | Kaposi's sarcoma-associated herpesvirus (KSHV), C12 portal dodecamer structure | | Descriptor: | Portal protein | | Authors: | Gong, D, Dai, X, Jih, J, Liu, Y.T, Bi, G.Q, Sun, R, Zhou, Z.H. | | Deposit date: | 2019-07-07 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | DNA-Packing Portal and Capsid-Associated Tegument Complexes in the Tumor Herpesvirus KSHV.
Cell, 178, 2019
|
|
2POY
 
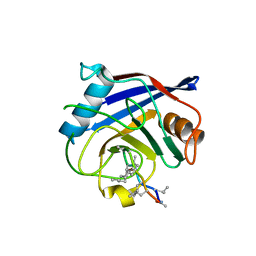 | | Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_4120 in complex with cyclosporin A | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE | | Authors: | Wernimont, A.K, Lew, J, Hills, T, Kozieradzki, I, Lin, Y.H, Hassanali, A, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-27 | | Release date: | 2007-05-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cryptosporidium Parvum Cyclophilin Type Peptidyl-Prolyl Cis-Trans Isomerase Cgd2_4120 in Complex with Cyclosporin A.
To be Published
|
|
6PKK
 
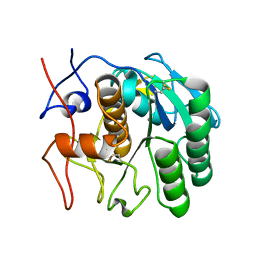 | | MicroED structure of proteinase K from an uncoated, single lamella at 2.18A resolution (#5) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.176 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
3C0Y
 
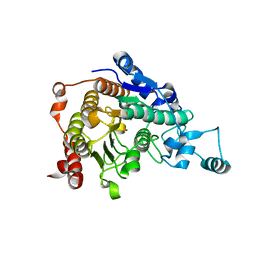 | | Crystal structure of catalytic domain of human histone deacetylase HDAC7 | | Descriptor: | Histone deacetylase 7a, POTASSIUM ION, ZINC ION | | Authors: | Min, J.R, Schuetz, A, Allali-Hassani, A, Loppnau, P, Kwiatkowski, N.P, Mazitschek, R, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-21 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human HDAC7 harbors a class IIa histone deacetylase-specific zinc binding motif and cryptic deacetylase activity.
J.Biol.Chem., 283, 2008
|
|
6PKP
 
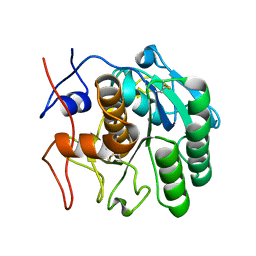 | | MicroED structure of proteinase K from a platinum-coated, polished, single lamella at 1.91A resolution (#10) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.91 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
2PPI
 
 | | Structure of the BTB (Tramtrack and Bric a brac) domain of human Gigaxonin | | Descriptor: | Gigaxonin | | Authors: | Amos, A, Turnbull, A.P, Tickle, J, Keates, T, Bullock, A, Savitsky, P, Burgess-Brown, N, Debreczeni, J.E, Ugochukwu, E, Umeano, C, Pike, A.C.W, Papagrigoriou, E, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-30 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the BTB (Tramtrack and Bric a brac) domain of human Gigaxonin.
To be Published
|
|
6PL0
 
 | | Crystal structure of the dark-adapted full-length bacteriophytochrome XccBphP from Xanthomonas campestris in the Pr state bound to BV chromophore | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Otero, L.H, Sirigu, S, Klinke, S, Goldbaum, F, Chavas, L, Rinaldi, J, Bonomi, H.R. | | Deposit date: | 2019-06-30 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structural basis for the Pr-Pfr long-range signaling mechanism of a full-length bacterial phytochrome at the atomic level.
Sci Adv, 7, 2021
|
|
4TU6
 
 | | Crystal structure of apo ATAD2A bromodomain with N1064 alternate conformation | | Descriptor: | ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-23 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
2XZE
 
 | |
2PWT
 
 | | Crystal structure of the bacterial ribosomal decoding site complexed with aminoglycoside containing the L-HABA group | | Descriptor: | 22-mer of the ribosomal decoding site, DOUBLY FUNCTIONALIZED PAROMOMYCIN PM-II-162 | | Authors: | Kondo, J, Pachamuthu, K, Francois, B, Szychowski, J, Hanessian, S, Westhof, E. | | Deposit date: | 2007-05-13 | | Release date: | 2007-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Bacterial Ribosomal Decoding Site Complexed with a Synthetic Doubly Functionalized Paromomycin Derivative: a New Specific Binding Mode to an A-Minor Motif Enhances in vitro Antibacterial Activity
Chemmedchem, 2, 2007
|
|
4I48
 
 | | Structure of HLA-A68 complexed with an HIV Env derived peptide | | Descriptor: | 9-mer peptide from Envelope glycoprotein gp160, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Niu, L, Cheng, H, Zhang, S, Tan, S, Zhang, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2012-11-27 | | Release date: | 2013-10-02 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Structural basis for the differential classification of HLA-A*6802 and HLA-A*6801 into the A2 and A3 supertypes
Mol.Immunol., 55, 2013
|
|
2PSB
 
 | | Crystal structure of YerB protein from Bacillus subtilis. NorthEast Structural Genomics target SR586 | | Descriptor: | YerB protein | | Authors: | Seetharaman, J, Chen, Y, Forouhar, F, Wang, D, Fang, Y, Cunningham, K, Ma, L.-C, Xia, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-05-04 | | Release date: | 2007-05-15 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of YerB protein from Bacillus subtilis.
To be Published
|
|
