7K1Q
 
 | |
7RS4
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with DMERI-8 | | Descriptor: | (2E)-3-{4-[(1E)-2-(2-chloro-4-fluorophenyl)-1-(2H-indazol-5-yl)but-1-en-1-yl]phenyl}prop-2-enoic acid, CHLORIDE ION, Estrogen receptor | | Authors: | Min, J, Nwachukwu, J.C, Min, C.K, Njeri, J.W, Srinivasan, S, Rangarajan, E.S, Nettles, C.C, Yan, S, Houtman, R, Griffin, P.R, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Dual-mechanism estrogen receptor inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RS2
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with DMERI-23 | | Descriptor: | (2E)-3-(4-{[(1S,2R,4S,5S,6S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]heptane-2-sulfonyl](2,2,2-trifluoroethyl)amino}phenyl)prop-2-enoic acid, Estrogen receptor | | Authors: | Min, J, Nwachukwu, J.C, Min, C.K, Njeri, J.W, Srinivasan, S, Rangarajan, E.S, Nettles, C.C, Yan, S, Houtman, R, Griffin, P.R, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Dual-mechanism estrogen receptor inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RRY
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with DMERI-20 | | Descriptor: | (1S,2R,4S,5S,6S)-5,6-bis(4-hydroxyphenyl)-N-{4-[3-(piperidin-1-yl)propoxy]phenyl}-N-(2,2,2-trifluoroethyl)-7-oxabicyclo[2.2.1]heptane-2-sulfonamide, CHLORIDE ION, Estrogen receptor | | Authors: | Min, J, Nwachukwu, J.C, Min, C.K, Njeri, J.W, Srinivasan, S, Rangarajan, E.S, Nettles, C.C, Yan, S, Houtman, R, Griffin, P.R, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Dual-mechanism estrogen receptor inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RS1
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with DMERI-21 | | Descriptor: | Estrogen receptor, methyl 3-(4-{[(1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonyl](2,2,2-trifluoroethyl)amino}phenyl)prop-2-enoate | | Authors: | Min, J, Nwachukwu, J.C, Min, C.K, Njeri, J.W, Srinivasan, S, Rangarajan, E.S, Nettles, C.C, Yan, S, Houtman, R, Griffin, P.R, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Dual-mechanism estrogen receptor inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RS3
 
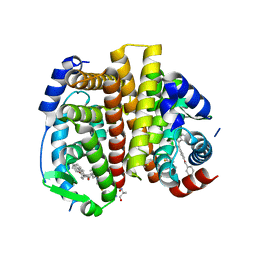 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with DMERI-29 | | Descriptor: | (1S,2R,4S)-6-[4-(benzyloxy)phenyl]-5-(4-hydroxyphenyl)-N-(4-methoxyphenyl)-N-(2,2,2-trifluoroethyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, CYSTEINE, Estrogen receptor, ... | | Authors: | Min, J, Nwachukwu, J.C, Min, C.K, Njeri, J.W, Srinivasan, S, Rangarajan, E.S, Nettles, C.C, Yan, S, Houtman, R, Griffin, P.R, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Dual-mechanism estrogen receptor inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RSQ
 
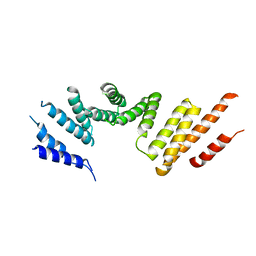 | | Cryo-EM structure of KIFBP core | | Descriptor: | KIF-binding protein | | Authors: | Solon, A.L, Tan, Z, Schutt, K.L, Jepsen, L, Haynes, S.E, Nesvizhskii, A.I, Sept, D, Stumpff, J, Ohi, R, Cianfrocco, M.A. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Kinesin-binding protein remodels the kinesin motor to prevent microtubule binding.
Sci Adv, 7, 2021
|
|
7RRZ
 
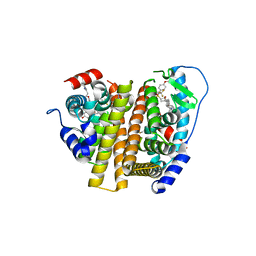 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with DMERI-30 | | Descriptor: | (1S,2R,4S,5R,6S)-5-(4-hydroxyphenyl)-N-(4-methoxyphenyl)-6-{4-[3-(piperidin-1-yl)propoxy]phenyl}-N-(2,2,2-trifluoroethyl)-7-oxabicyclo[2.2.1]heptane-2-sulfonamide, Estrogen receptor | | Authors: | Min, J, Nwachukwu, J.C, Min, C.K, Njeri, J.W, Srinivasan, S, Rangarajan, E.S, Nettles, C.C, Yan, S, Houtman, R, Griffin, P.R, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Dual-mechanism estrogen receptor inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3C0K
 
 | |
7RS0
 
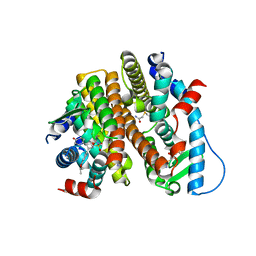 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with DMERI-18 | | Descriptor: | (1R,2S,4R,5R,6R)-5-(4-hydroxyphenyl)-N-(4-methoxyphenyl)-6-(4-propoxyphenyl)-N-(2,2,2-trifluoroethyl)-7-oxabicyclo[2.2.1]heptane-2-sulfonamide, Estrogen receptor | | Authors: | Min, J, Nwachukwu, J.C, Min, C.K, Njeri, J.W, Srinivasan, S, Rangarajan, E.S, Nettles, C.C, Yan, S, Houtman, R, Griffin, P.R, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Dual-mechanism estrogen receptor inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RFK
 
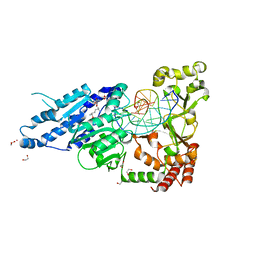 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor Sinefungin | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Cheng, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Repurposing epigenetic inhibitors to target the Clostridioides difficile- specific DNA adenine methyltransferase and sporulation regulator CamA.
Epigenetics, 17, 2022
|
|
7RFL
 
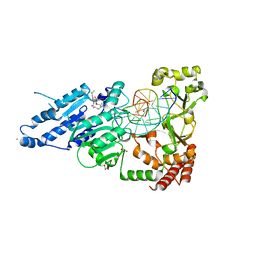 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor SGC0946 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, DNA Strand 1, ... | | Authors: | Horton, J.R, Cheng, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Repurposing epigenetic inhibitors to target the Clostridioides difficile- specific DNA adenine methyltransferase and sporulation regulator CamA.
Epigenetics, 17, 2022
|
|
7RM5
 
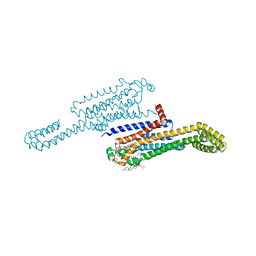 | | MicroED structure of the human adenosine receptor at 2.8A | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a/Soluble cytochrome b562 chimera, CHOLESTEROL, ... | | Authors: | Martynowycz, M.W, Shiriaeva, A, Ge, X, Hattne, J, Nannenga, B.L, Cherezov, V, Gonen, T. | | Deposit date: | 2021-07-26 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.79 Å) | | Cite: | MicroED structure of the human adenosine receptor determined from a single nanocrystal in LCP.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1ZK1
 
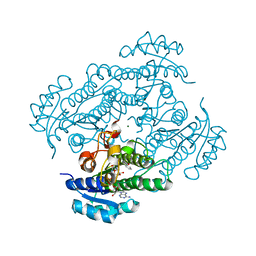 | | Structure of R-specific alcohol dehydrogenase (mutant G37D) from Lactobacillus brevis in complex with phenylethanol and NAD | | Descriptor: | 1-PHENYLETHANONE, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Schlieben, N.H, Niefind, K, Muller, J, Riebel, B, Hummel, W, Schomburg, D. | | Deposit date: | 2005-05-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Atomic Resolution Structures of R-specific Alcohol Dehydrogenase from Lactobacillus brevis Provide the Structural Bases of its Substrate and Cosubstrate Specificity
J.Mol.Biol., 349, 2005
|
|
7JMC
 
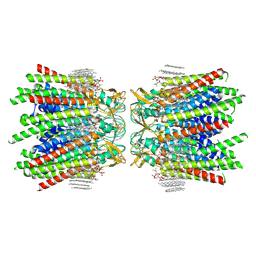 | | Sheep Connexin-50 at 2.5 angstroms resolution, Lipid Class 3 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
2VDL
 
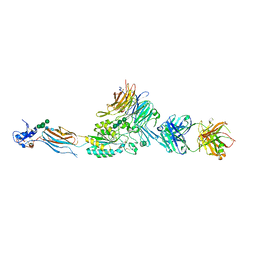 | | Re-refinement of Integrin AlphaIIbBeta3 Headpiece | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Springer, T.A, Zhu, J, Xiao, T. | | Deposit date: | 2007-10-10 | | Release date: | 2008-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis for Distinctive Recognition of Fibrinogen Gammac Peptide by the Platelet Integrin Alphaiibbeta3.
J.Cell Biol., 182, 2008
|
|
7RFM
 
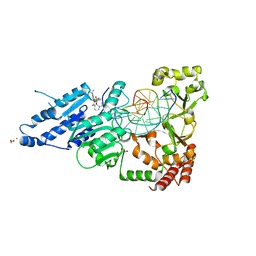 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor EPZ004777 | | Descriptor: | 1,2-ETHANEDIOL, 7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, DNA Strand 1, ... | | Authors: | Horton, J.R, Cheng, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Repurposing epigenetic inhibitors to target the Clostridioides difficile- specific DNA adenine methyltransferase and sporulation regulator CamA.
Epigenetics, 17, 2022
|
|
7RFN
 
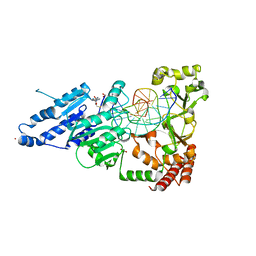 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor SGC8158 | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-(4-{[(4'-chloro[1,1'-biphenyl]-3-yl)methyl]amino}butyl)-5'-thioadenosine, DNA Strand 1, ... | | Authors: | Horton, J.R, Cheng, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Repurposing epigenetic inhibitors to target the Clostridioides difficile- specific DNA adenine methyltransferase and sporulation regulator CamA.
Epigenetics, 17, 2022
|
|
6YWN
 
 | | CutA in complex with CMPCPP | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, CALCIUM ION, CutA | | Authors: | Malik, D, Kobylecki, K, Krawczyk, P, Poznanski, J, Jakielaszek, A, Napiorkowska, A, Dziembowski, A, Tomecki, R, Nowotny, M. | | Deposit date: | 2020-04-29 | | Release date: | 2020-08-05 | | Last modified: | 2020-09-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and mechanism of CutA, RNA nucleotidyl transferase with an unusual preference for cytosine.
Nucleic Acids Res., 48, 2020
|
|
7REI
 
 | | The crystal structure of nickel bound human ADO C18S C239S variant | | Descriptor: | 2-aminoethanethiol dioxygenase, GLYCEROL, NICKEL (II) ION | | Authors: | Wang, Y, Shin, I, Li, J, Liu, A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of human cysteamine dioxygenase provides a structural rationale for its function as an oxygen sensor.
J.Biol.Chem., 297, 2021
|
|
5JBE
 
 | | 4,6-alpha-glucanotransferase GTFB from Lactobacillus reuteri 121 complexed with an isomalto-maltopentasaccharide | | Descriptor: | ACETATE ION, CALCIUM ION, Inactive glucansucrase, ... | | Authors: | Pijning, T, Dijkstra, B.W, Bai, Y, Gangoiti-Munecas, J, Dijkhuizen, L. | | Deposit date: | 2016-04-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of 4,6-alpha-Glucanotransferase Supports Diet-Driven Evolution of GH70 Enzymes from alpha-Amylases in Oral Bacteria.
Structure, 25, 2017
|
|
7RRX
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with DMERI-19 | | Descriptor: | (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-N-{4-[2-(piperidin-1-yl)ethoxy]phenyl}-N-(2,2,2-trifluoroethyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, CHLORIDE ION, Estrogen receptor | | Authors: | Min, J, Nwachukwu, J.C, Min, C.K, Njeri, J.W, Srinivasan, S, Rangarajan, E.S, Nettles, C.C, Yan, S, Houtman, R, Griffin, P.R, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Dual-mechanism estrogen receptor inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6Z22
 
 | |
7RS8
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with DMERI-16 | | Descriptor: | (1R,2S,4R)-5-(4-hydroxyphenyl)-N-(4-methoxyphenyl)-6-{4-[2-(piperidin-1-yl)ethoxy]phenyl}-N-(2,2,2-trifluoroethyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor | | Authors: | Min, J, Nwachukwu, J.C, Min, C.K, Njeri, J.W, Srinivasan, S, Rangarajan, E.S, Nettles, C.C, Yan, S, Houtman, R, Griffin, P.R, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Dual-mechanism estrogen receptor inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6YVH
 
 | | CWC22-CWC27-EIF4A3 Complex | | Descriptor: | Eukaryotic initiation factor 4A-III, Pre-mRNA-splicing factor CWC22 homolog, Spliceosome-associated protein CWC27 homolog | | Authors: | Basquin, J, Busetto, V, LeHir, H, Conti, E. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structural and functional insights into CWC27/CWC22 heterodimer linking the exon junction complex to spliceosomes.
Nucleic Acids Res., 48, 2020
|
|
