6LPU
 
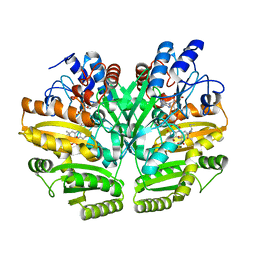 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in complex with L-2-hydroxyglutarate (L-2-HG) | | Descriptor: | (2S)-2-HYDROXYPENTANEDIOIC ACID, D-2-hydroxyglutarate dehydrogenase, mitochondrial, ... | | Authors: | Yang, J, Zhu, H, Ding, J. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.923 Å) | | Cite: | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
3AVC
 
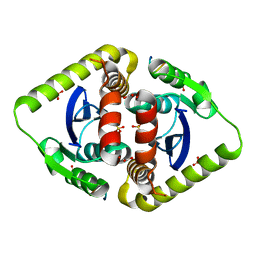 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-02 | | Release date: | 2012-01-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
3AVK
 
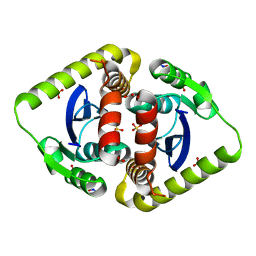 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-05 | | Release date: | 2012-01-18 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
5AR8
 
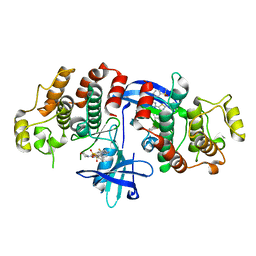 | | RIP2 Kinase Catalytic Domain (1 - 310) complex with Biphenylsulfonamide | | Descriptor: | 2,6-bis(fluoranyl)-N-[3-[5-[2-[(3-methylsulfonylphenyl)amino]pyrimidin-4-yl]-2-morpholin-4-yl-1,3-thiazol-4-yl]phenyl]benzenesulfonamide, RECEPTOR-INTERACTING SERINE/THREONINE-PROTEIN KINASE 2 | | Authors: | Charnley, A.K, Convery, M.A, Lakdawala Shah, A, Jones, E, Hardwicke, P, Bridges, A, Votta, B.J, Gough, P.J, Marquis, R.W, Bertin, J, Casillas, L. | | Deposit date: | 2015-09-24 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal Structures of Human Rip2 Kinase Catalytic Domain Complexed with ATP-Competitive Inhibitors: Foundations for Understanding Inhibitor Selectivity.
Bioorg.Med.Chem., 23, 2015
|
|
6VW0
 
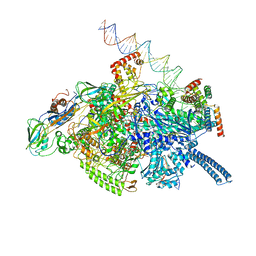 | | Mycobacterium tuberculosis RNAP S456L mutant open promoter complex | | Descriptor: | DNA (65-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Lilic, M, Boyaci, H, Chen, J, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3AVE
 
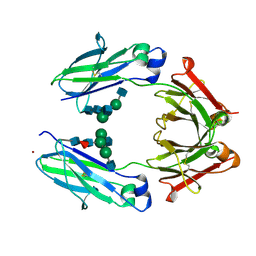 | | Crystal Structure of the Fucosylated Fc Fragment from Human Immunoglobulin G1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region, ZINC ION | | Authors: | Matsumiya, S, Yamaguchi, Y, Saito, J, Nagano, M, Sasakawa, H, Otaki, S, Satoh, M, Shitara, K, Kato, K. | | Deposit date: | 2011-03-04 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Corrigendum to "Structural Comparison of Fucosylated and Nonfucosylated Fc Fragments of Human Immunoglobulin G1" [J. Mol. Biol. 386/3 (2007) 767-779]
J.Mol.Biol., 408, 2011
|
|
4RL1
 
 | |
7EMA
 
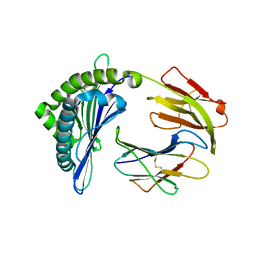 | | Mooring Stone-Like Arg114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I | | Descriptor: | Beta-2-microglobulin, Leucocyte antigen, TYR-SER-SER-ASP-VAL-THR-THR-LEU-VAL | | Authors: | Yue, C, Xiang, W, Huang, X, Sun, Y, Xiao, J, Liu, K, Sun, Z, Qiao, P, Li, H, Gan, J, Ba, L, Chai, Y, Qi, J, Liu, P, Qi, P, Zhao, Y, Li, Y, Qiu, H.J, Gao, G.F, Gao, G, Liu, W.J. | | Deposit date: | 2021-04-13 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mooring Stone-Like Arg 114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I.
J.Virol., 96, 2022
|
|
7EMC
 
 | | Mooring Stone-Like Arg114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I | | Descriptor: | ALA-THR-GLU-ILE-ARG-GLU-LEU-LEU-VAL, Beta-2-microglobulin, Leucocyte antigen | | Authors: | Yue, C, Xiang, W, Huang, X, Sun, Y, Xiao, J, Liu, K, Sun, Z, Qiao, P, Li, H, Gan, J, Ba, L, Chai, Y, Qi, J, Liu, P, Qi, P, Zhao, Y, Li, Y, Qiu, H.J, Gao, G.F, Gao, G, Liu, W.J. | | Deposit date: | 2021-04-13 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mooring Stone-Like Arg 114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I.
J.Virol., 96, 2022
|
|
7EMB
 
 | | Mooring Stone-Like Arg114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I | | Descriptor: | ALA-ALA-ALA-ILE-GLU-GLU-GLU-ASP-ILE, Beta-2-microglobulin, Leucocyte antigen | | Authors: | Yue, C, Xiang, W, Huang, X, Sun, Y, Xiao, J, Liu, K, Sun, Z, Qiao, P, Li, H, Gan, J, Ba, L, Chai, Y, Qi, J, Liu, P, Qi, P, Zhao, Y, Li, Y, Qiu, H.J, Gao, G.F, Gao, G, Liu, W.J. | | Deposit date: | 2021-04-13 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Mooring Stone-Like Arg 114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I.
J.Virol., 96, 2022
|
|
7EM9
 
 | | Mooring Stone-Like Arg114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I | | Descriptor: | Beta-2-microglobulin, Leucocyte antigen, SER-LEU-ASP-GLU-TYR-SER-SER-ASP-VAL | | Authors: | Yue, C, Xiang, W, Huang, X, Sun, Y, Xiao, J, Liu, K, Sun, Z, Qiao, P, Li, H, Gan, J, Ba, L, Chai, Y, Qi, J, Liu, P, Qi, P, Zhao, Y, Li, Y, Qiu, H.J, Gao, G.F, Gao, G, Liu, W.J. | | Deposit date: | 2021-04-13 | | Release date: | 2021-12-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mooring Stone-Like Arg 114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I.
J.Virol., 96, 2022
|
|
7EMD
 
 | | Mooring Stone-Like Arg114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I | | Descriptor: | Beta-2-microglobulin, Leucocyte antigen, TYR-GLY-ASP-PHE-PHE-HIS-ASP-MET-VAL | | Authors: | Yue, C, Xiang, W, Huang, X, Sun, Y, Xiao, J, Liu, K, Sun, Z, Qiao, P, Li, H, Gan, J, Ba, L, Chai, Y, Qi, J, Liu, P, Qi, P, Zhao, Y, Li, Y, Qiu, H.J, Gao, G.F, Gao, G, Liu, W.J. | | Deposit date: | 2021-04-13 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mooring Stone-Like Arg 114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I.
J.Virol., 96, 2022
|
|
6NQ2
 
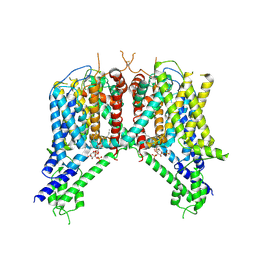 | | Cryo-EM structure of human TPC2 channel in the ligand-bound closed state | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, Two pore calcium channel protein 2 | | Authors: | She, J, Zeng, W, Guo, J, Chen, Q, Bai, X, Jiang, Y. | | Deposit date: | 2019-01-18 | | Release date: | 2019-03-27 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural mechanisms of phospholipid activation of the human TPC2 channel.
Elife, 8, 2019
|
|
5AML
 
 | |
2ERX
 
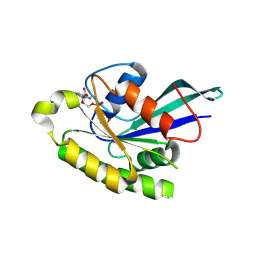 | | Crystal Structure of DiRas2 in Complex With GDP and Inorganic Phosphate | | Descriptor: | GTP-binding protein Di-Ras2, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Papagrigoriou, E, Yang, X, Elkins, J, Niesen, F.E, Burgess, N, Salah, E, Fedorov, O, Ball, L.J, von Delft, F, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, Doyle, D. | | Deposit date: | 2005-10-25 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of DiRas2
To be Published
|
|
3AVM
 
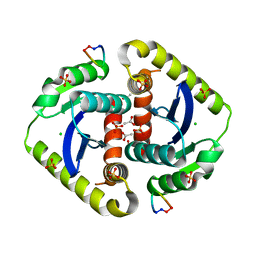 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-05 | | Release date: | 2012-01-18 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
5TZY
 
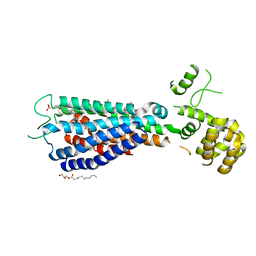 | | GPR40 in complex with AgoPAM AP8 and partial agonist MK-8666 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S,3R)-3-cyclopropyl-3-[(2R)-2-(1-{(1S)-1-[5-fluoro-2-(trifluoromethoxy)phenyl]ethyl}piperidin-4-yl)-3,4-dihydro-2H-1-benzopyran-7-yl]-2-methylpropanoic acid, (5aR,6S,6aS)-3-({2',6'-dimethyl-4'-[3-(methylsulfonyl)propoxy][1,1'-biphenyl]-3-yl}methoxy)-5,5a,6,6a-tetrahydrocyclopropa[4,5]cyclopenta[1,2-c]pyridine-6-carboxylic acid, ... | | Authors: | Lu, J, Byrne, N, Patel, S, Sharma, S, Soisson, S.M. | | Deposit date: | 2016-11-22 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6W4Q
 
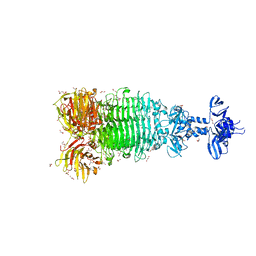 | | Crystal structure of full-length tailspike protein 2 (TSP2, ORF211) ) from Escherichia coli O157:H7 bacteriophage CAB120 | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, CHLORIDE ION, ... | | Authors: | Greenfield, J, Herzberg, O. | | Deposit date: | 2020-03-11 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of bacteriophage CBA120 ORF211 (TSP2), the determinant of phage specificity towards E. coli O157:H7.
Sci Rep, 10, 2020
|
|
6W89
 
 | | Structure of DNMT3A (R882H) in complex with CGA DNA | | Descriptor: | CGA DNA (25-MER), CITRIC ACID, DNA (cytosine-5)-methyltransferase 3-like, ... | | Authors: | Anteneh, H, Song, J. | | Deposit date: | 2020-03-20 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Structural basis for impairment of DNA methylation by the DNMT3A R882H mutation.
Nat Commun, 11, 2020
|
|
1STZ
 
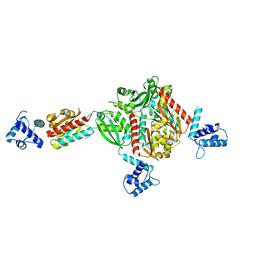 | | Crystal structure of a hypothetical protein at 2.2 A resolution | | Descriptor: | Heat-inducible transcription repressor hrcA homolog | | Authors: | Liu, J, Adams, P.D, Shin, D.-H, Huang, C, Yokota, H, Jancarik, J, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-03-25 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a heat-inducible transcriptional repressor HrcA from Thermotoga maritima: structural insight into DNA binding and dimerization.
J.Mol.Biol., 350, 2005
|
|
5DP9
 
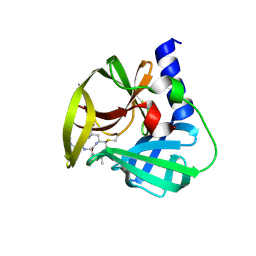 | | Crystal Structure of EV71 3C Proteinase in complex with compound 9 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-[(N-{[(cyclobutylmethyl)amino](oxo)acetyl}-L-phenylalanyl)amino]-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
3AVG
 
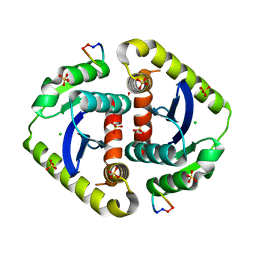 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-05 | | Release date: | 2012-01-18 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
2F9C
 
 | | Crystal structure of YDCK from Salmonella cholerae. NESG target SCR6 | | Descriptor: | CESIUM ION, Hypothetical protein YDCK | | Authors: | Benach, J, Chen, Y, Vorobiev, S.M, Seetharaman, J, Janjua, H, Cooper, B, Acton, X.T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-05 | | Release date: | 2005-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of YDCK from Salmonella cholerae NESG target SCR6.
To be Published
|
|
3FS6
 
 | | Correlations of Inhibitor Kinetics for Pneumocystis jirovecii and Human Dihydrofolate Reductase with Structural Data for Human Active Site Mutant Enzyme Complexes | | Descriptor: | 2,4-DIAMINO-5-[2-METHOXY-5-(4-CARBOXYBUTYLOXY)BENZYL]PYRIMIDINE, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Pace, J, Makin, J, Piraino, J, Queener, S.F, Rosowsky, A. | | Deposit date: | 2009-01-09 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Correlations of Inhibitor Kinetics for Pneumocystis jirovecii and Human Dihydrofolate Reductase with Structural Data for Human Active Site Mutant Enzyme Complexes.
Biochemistry, 48, 2009
|
|
6NQ1
 
 | | Cryo-EM structure of human TPC2 channel in the apo state | | Descriptor: | Two pore calcium channel protein 2 | | Authors: | She, J, Zeng, W, Guo, J, Chen, Q, Bai, X, Jiang, Y. | | Deposit date: | 2019-01-18 | | Release date: | 2019-03-27 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural mechanisms of phospholipid activation of the human TPC2 channel.
Elife, 8, 2019
|
|
