3FJF
 
 | |
3F5M
 
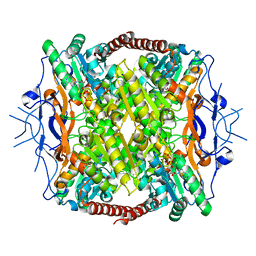 | | Crystal Structure of ATP-Bound Phosphofructokinase from Trypanosoma brucei | | Descriptor: | 6-phospho-1-fructokinase (ATP-dependent phosphofructokinase), ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | McNae, I.W, Martinez-Oyanedel, J, Keillor, J.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2008-11-04 | | Release date: | 2008-11-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of ATP-bound phosphofructokinase from Trypanosoma brucei reveals conformational transitions different from those of other phosphofructokinases.
J.Mol.Biol., 385, 2009
|
|
3F6J
 
 | | F17a-G lectin domain with bound GlcNAc(beta1-3)Gal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-methyl beta-D-galactopyranoside, F17a-G | | Authors: | Buts, L, de Boer, A, Olsson, J.D.M, Jonckheere, W, De Kerpel, M, De Genst, E, Guerardel, Y, Willaert, R, Wyns, L, Wuhrer, M, Oscarson, S, De Greve, H, Bouckaert, J. | | Deposit date: | 2008-11-06 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Sampling of Glycan Interaction Profiles Reveals Mucosal Receptors for Fimbrial Adhesins of Enterotoxigenic Escherichia coli
Biology, 2, 2013
|
|
6BPE
 
 | | Plasmodium vivax reticulocyte binding protein 2b (PvRBP2b) bound to monoclonal antibody 6H1 | | Descriptor: | Monoclonal antibody 6H1 Fab heavy chain, Monoclonal antibody 6H1 Fab light chain, Reticulocyte binding protein 2, ... | | Authors: | Gruszczyk, J, Chan, L.J, Tham, W.H. | | Deposit date: | 2017-11-22 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Cryo-EM structure of an essential Plasmodium vivax invasion complex.
Nature, 559, 2018
|
|
6FUJ
 
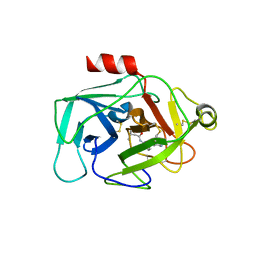 | | Complement factor D in complex with the inhibitor N-(3'-(aminomethyl)-[1,1'-biphenyl]-3-yl)-3-methylbutanamide | | Descriptor: | Complement factor D, ~{N}-[3-[3-(aminomethyl)phenyl]phenyl]-3-methyl-butanamide | | Authors: | Mac Sweeney, A, Ostermann, N, Vulpetti, A, Maibaum, J, Erbel, P, Lorthiois, E, Yoon, T, Randl, S, Ruedisser, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
1GHK
 
 | | SOLUTION STRUCTURE OF THE LIPOYL DOMAIN OF THE 2-OXOGLUTARATE DEHYDROGENASE COMPLEX FROM AZOTOBACTER VINELAND II, NMR, 25 STRUCTURES | | Descriptor: | E2, THE DIHYDROLIPOAMIDE SUCCINYLTRANSFERASE COMPONENT OF 2-OXOGLUTARATE DEHYDROGENASE COMPLEX | | Authors: | Berg, A, Vervoort, J, De Kok, A. | | Deposit date: | 1996-01-16 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the lipoyl domain of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii.
J.Mol.Biol., 261, 1996
|
|
1GHJ
 
 | | SOLUTION STRUCTURE OF THE LIPOYL DOMAIN OF THE 2-OXOGLUTARATE DEHYDROGENASE COMPLEX FROM AZOTOBACTER VINELAND II, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | E2, THE DIHYDROLIPOAMIDE SUCCINYLTRANSFERASE COMPONENT OF 2-OXOGLUTARATE DEHYDROGENASE COMPLEX | | Authors: | Berg, A, Vervoort, J, De Kok, A. | | Deposit date: | 1996-01-16 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the lipoyl domain of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii.
J.Mol.Biol., 261, 1996
|
|
3FN3
 
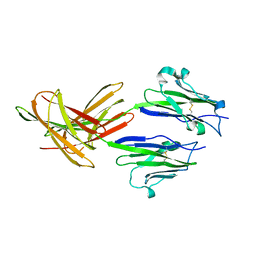 | | Dimeric Structure of PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Chen, Y, Gao, F, Liu, P, Chu, F, Qi, J, Gao, G.F. | | Deposit date: | 2008-12-23 | | Release date: | 2009-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A dimeric structure of PD-L1: functional units or evolutionary relics?
Protein Cell, 1, 2010
|
|
6C5W
 
 | | Crystal structure of the mitochondrial calcium uniporter | | Descriptor: | CALCIUM ION, calcium uniporter, nanobody | | Authors: | Fan, C, Fan, M, Fastman, N, Zhang, J, Feng, L. | | Deposit date: | 2018-01-17 | | Release date: | 2018-07-11 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (3.10010242 Å) | | Cite: | X-ray and cryo-EM structures of the mitochondrial calcium uniporter.
Nature, 559, 2018
|
|
3FO5
 
 | | Human START domain of Acyl-coenzyme A thioesterase 11 (ACOT11) | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Siponen, M.I, Lehtio, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Thorsell, A.G, Tresaugues, L, Van-Den-Berg, S, Weigelt, J, Welin, M, Wikstrom, M, Wisniewska, M, Shueler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-27 | | Release date: | 2009-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Comparative structural analysis of lipid binding START domains.
Plos One, 6, 2011
|
|
3E0C
 
 | | Crystal Structure of DNA Damage-Binding protein 1(DDB1) | | Descriptor: | DNA damage-binding protein 1 | | Authors: | Amaya, M.F, Xu, L, Hao, H, Bountra, C, Wickstroem, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-07-31 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure and function of WD40 domain proteins.
Protein Cell, 2, 2011
|
|
3DXD
 
 | |
3E1E
 
 | | Crystal structure of a Thioesterase family protein from Silicibacter pomeroyi. NorthEast Structural Genomics target SiR180A | | Descriptor: | Thioesterase family protein | | Authors: | Seetharaman, J, Abashidze, M, Wang, H, Janjua, H, Foote, E.L, Xiao, R, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-08-04 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a Thioesterase family protein from Silicibacter pomeroyi. NorthEast Structural Genomics target SiR180A
To be Published
|
|
6DRW
 
 | |
3DUZ
 
 | | Crystal structure of the postfusion form of baculovirus fusion protein GP64 | | Descriptor: | MERCURY (II) ION, Major envelope glycoprotein | | Authors: | Kadlec, J, Loureiro, S, Abrescia, N.G.A, Jones, I.M, Stuart, D.I. | | Deposit date: | 2008-07-18 | | Release date: | 2008-09-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines.
Nat.Struct.Mol.Biol., 15, 2008
|
|
6DV6
 
 | | Structure of the Salmonella SPI-1 type III secretion injectisome secretin InvG (residues 176-end) in the open gate state | | Descriptor: | Protein InvG | | Authors: | Hu, J, Worrall, L.J, Vuckovic, M, Atkinson, C.E, Strynadka, N.C.J. | | Deposit date: | 2018-06-22 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM analysis of the T3S injectisome reveals the structure of the needle and open secretin.
Nat Commun, 9, 2018
|
|
3E1R
 
 | | Midbody targeting of the ESCRT machinery by a non-canonical coiled-coil in CEP55 | | Descriptor: | Centrosomal protein of 55 kDa, Programmed cell death 6-interacting protein | | Authors: | Lee, H.H, Elia, N, Ghirlando, R, Lippincott-Schwartz, J, Hurley, J.H. | | Deposit date: | 2008-08-04 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55.
Science, 322, 2008
|
|
6DD5
 
 | | Crystal Structure of the Cas6 Domain of Marinomonas mediterranea MMB-1 Cas6-RT-Cas1 Fusion Protein | | Descriptor: | GLYCEROL, MMB-1 Cas6 Fused to Maltose Binding Protein,CRISPR-associated endonuclease Cas1, SULFATE ION, ... | | Authors: | Stamos, J.L, Mohr, G, Silas, S, Makarova, K.S, Markham, L.M, Yao, J, Lucas-Elio, P, Sanchez-Amat, A, Fire, A.Z, Koonin, E.V, Lambowitz, A.M. | | Deposit date: | 2018-05-09 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Reverse Transcriptase-Cas1 Fusion Protein Contains a Cas6 Domain Required for Both CRISPR RNA Biogenesis and RNA Spacer Acquisition.
Mol. Cell, 72, 2018
|
|
6CAU
 
 | | UDP-N-acetylmuramate--alanine ligase from Acinetobacter baumannii AB5075-UW with AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, UDP-N-acetylmuramate--L-alanine ligase | | Authors: | Horanyi, P.S, Abendroth, J, Lorimer, D.D, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-01-31 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | UDP-N-acetylmuramate--alanine ligase from Acinetobacter baumannii AB5075-UW with AMPPNP
To be Published
|
|
6DDY
 
 | | Crystal structure of the double mutant (D52N/K359Q) of NT5C2-537X in the active state, Northeast Structural Genomics Target | | Descriptor: | Cytosolic purine 5'-nucleotidase, GLYCEROL, PHOSPHATE ION | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
2K3Q
 
 | |
6DGP
 
 | |
3E28
 
 | |
5OGR
 
 | | Structure of cathepsin B1 from Schistosoma mansoni in complex with WRR286 inhibitor | | Descriptor: | 3-[[N-[4-METHYL-PIPERAZINYL]CARBONYL]-PHENYLALANINYL-AMINO]-5-PHENYL-PENTANE-1-SULFONIC ACID BENZYLOXY-AMIDE, ACETATE ION, Cathepsin B-like peptidase (C01 family) | | Authors: | Jilkova, A, Rezacova, P, Brynda, J, Mares, M. | | Deposit date: | 2017-07-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Druggable Hot Spots in the Schistosomiasis Cathepsin B1 Target Identified by Functional and Binding Mode Analysis of Potent Vinyl Sulfone Inhibitors.
Acs Infect Dis., 2020
|
|
6BIE
 
 | |
