1S2H
 
 | | The Mad2 spindle checkpoint protein possesses two distinct natively folded states | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2A | | Authors: | Luo, X, Tang, Z, Xia, G, Wassmann, K, Matsumoto, T, Rizo, J, Yu, H. | | Deposit date: | 2004-01-08 | | Release date: | 2004-03-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Mad2 spindle checkpoint protein has two distinct natively folded states.
Nat.Struct.Mol.Biol., 11, 2004
|
|
3HLR
 
 | | Donor strand complemented FaeG of F4ad fimbriae | | Descriptor: | K88 fimbrial protein AD | | Authors: | Van Molle, I, Moonens, K, Garcia-Pino, A, Buts, L, Bouckaert, J, De Greve, H. | | Deposit date: | 2009-05-28 | | Release date: | 2009-10-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and thermodynamic characterization of pre- and postpolymerization states in the F4 fimbrial subunit FaeG
J.Mol.Biol., 394, 2009
|
|
3VFC
 
 | | Crystal structure of enolase MSMEG_6132 (TARGET EFI-502282) from Mycobacterium smegmatis str. MC2 155 complexed with tartrate | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, L(+)-TARTARIC ACID, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-09 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of enolase MSMEG_6132 from Mycobacterium smegmatis
To be Published
|
|
3HLM
 
 | | Crystal Structure of Mouse Mitochondrial Aspartate Aminotransferase/Kynurenine Aminotransferase IV | | Descriptor: | Aspartate aminotransferase, mitochondrial, GLYCEROL | | Authors: | Han, Q, Robinson, H, Li, J. | | Deposit date: | 2009-05-27 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure, expression, and function of kynurenine aminotransferases in human and rodent brains.
Cell.Mol.Life Sci., 67, 2010
|
|
3HLT
 
 | | The crystal structure of human haloacid dehalogenase-like hydrolase domain containing 2 (HDHD2) | | Descriptor: | CHLORIDE ION, HDHD2, SODIUM ION, ... | | Authors: | Ugochukwu, E, Shafqat, N, Yue, W.W, Cocking, R, Bray, J.E, Muniz, J.R.C, Krojer, T, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-05-28 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of human haloacid dehalogenase-like hydrolase domain containing 2 (HDHD2)
To be Published
|
|
3HF7
 
 | | The Crystal Structure of a CBS-domain Pair with Bound AMP from Klebsiella pneumoniae to 2.75A | | Descriptor: | ADENOSINE MONOPHOSPHATE, uncharacterized CBS-domain protein | | Authors: | Stein, A.J, Nocek, B, Wu, R, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-11 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Crystal Structure of a CBS-domain Pair with Bound AMP from Klebsiella pneumoniae to 2.75A
To be Published
|
|
1S34
 
 | | Solution structure of residues 907-929 from Rous Sarcoma Virus | | Descriptor: | 5'-R(*GP*GP*GP*GP*AP*GP*UP*GP*GP*UP*UP*UP*GP*UP*AP*UP*CP*CP*UP*UP*CP*CP*C)-3' | | Authors: | Cabello-Villegas, J, Giles, K.E, Soto, A.M, Yu, P, Beemon, K.L, Wang, Y.X. | | Deposit date: | 2004-01-12 | | Release date: | 2004-08-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the pseudo-5' splice site of a retroviral splicing suppressor.
Rna, 10, 2004
|
|
3HLW
 
 | | CTX-M-9 S70G in complex with cefotaxime | | Descriptor: | (6R,7R)-3-(acetyloxymethyl)-7-[[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-methoxyimino-ethanoyl]amino]-8-oxo-5-thia-1-azabicy clo[4.2.0]oct-2-ene-2-carboxylic acid, CTX-M-9 extended-spectrum beta-lactamase | | Authors: | Delmas, J, Leyssne, D, Dubois, D, Vazeille, E, Robin, F, Bonnet, R. | | Deposit date: | 2009-05-28 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into substrate recognition and product expulsion in CTX-M enzymes.
J.Mol.Biol., 400, 2010
|
|
3HG6
 
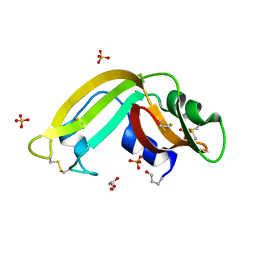 | | Crystal Structure of the Recombinant Onconase from Rana pipiens | | Descriptor: | GLYCEROL, Onconase, SULFATE ION | | Authors: | Camara-Artigas, A, Gavira, J.A, Casares-Atienza, S, Weininger, U, Balbach, J, Garcia-Mira, M.M. | | Deposit date: | 2009-05-13 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Three-state thermal unfolding of onconase.
Biophys.Chem., 159, 2011
|
|
3VI2
 
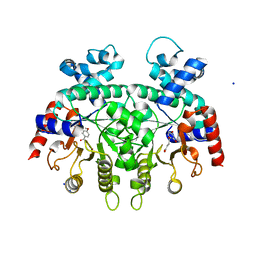 | | Crystal Structure Analysis of Plasmodium falciparum OMP Decarboxylase in complex with inhibitor HMOA | | Descriptor: | 4-(2-hydroxy-4-methoxyphenyl)-4-oxobutanoic acid, Orotidine 5'-phosphate decarboxylase, SODIUM ION | | Authors: | Takashima, Y, Mizohata, E, Krungkrai, S.R, Matsumura, H, Krungkrai, J, Horii, T, Inoue, T. | | Deposit date: | 2011-09-16 | | Release date: | 2012-08-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The in silico screening and X-ray structure analysis of the inhibitor complex of Plasmodium falciparum orotidine 5'-monophosphate decarboxylase
J.Biochem., 152, 2012
|
|
3HNQ
 
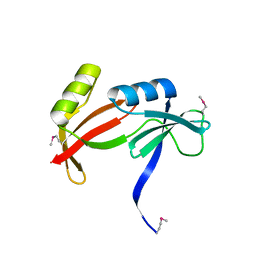 | | Crystal Structure of Virulence protein STM3117 from Salmonella typhimurium. Northeast Structural Genomics Consortium target id StR274 | | Descriptor: | Virulence protein STM3117 | | Authors: | Seetharaman, J, Su, M, Sahdev, S, Janjua, H, Xiao, R, Ciccosanti, C, Foote, E.L, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-05-31 | | Release date: | 2009-06-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Virulence protein STM3117 from Salmonella typhimurium. Northeast Structural Genomics Consortium target id StR274
To be Published
|
|
3VOB
 
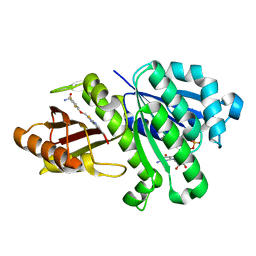 | | Staphylococcus aureus FtsZ with PC190723 | | Descriptor: | 3-[(6-chloro[1,3]thiazolo[5,4-b]pyridin-2-yl)methoxy]-2,6-difluorobenzamide, CALCIUM ION, Cell division protein FtsZ, ... | | Authors: | Yamane, J, Matsui, T, Mogi, N, Yamaguchi, H, Takemoto, H, Yao, M, Tanaka, I. | | Deposit date: | 2012-01-20 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structural reorganization of the bacterial cell-division protein FtsZ from Staphylococcus aureus
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3HOM
 
 | |
1SH2
 
 | | Crystal Structure of Norwalk Virus Polymerase (Metal-free, Centered Orthorhombic) | | Descriptor: | RNA Polymerase | | Authors: | Ng, K.K, Pendas-Franco, N, Rojo, J, Boga, J.A, Machin, A, Alonso, J.M, Parra, F. | | Deposit date: | 2004-02-24 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of norwalk virus polymerase reveals the carboxyl terminus in the active site cleft.
J.Biol.Chem., 279, 2004
|
|
1OMC
 
 | | SOLUTION STRUCTURE OF OMEGA-CONOTOXIN GVIA USING 2-D NMR SPECTROSCOPY AND RELAXATION MATRIX ANALYSIS | | Descriptor: | OMEGA-CONOTOXIN GVIA | | Authors: | Davis, J.H, Bradley, E.K, Miljanich, G.P, Nadasdi, L, Ramachandran, J, Basus, V.J. | | Deposit date: | 1993-04-28 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of omega-conotoxin GVIA using 2-D NMR spectroscopy and relaxation matrix analysis.
Biochemistry, 32, 1993
|
|
3VPI
 
 | |
1SGH
 
 | | Moesin FERM domain bound to EBP50 C-terminal peptide | | Descriptor: | Ezrin-radixin-moesin binding phosphoprotein 50, Moesin | | Authors: | Finnerty, C.M, Chambers, D, Ingraffea, J, Faber, H.R, Karplus, P.A, Bretscher, A. | | Deposit date: | 2004-02-23 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The EBP50-moesin interaction involves a binding site regulated by direct masking on the FERM domain
J.Cell.Sci., 117, 2004
|
|
3HQY
 
 | | Discovery of novel inhibitors of PDE10A | | Descriptor: | 2-({4-[4-(pyridin-4-ylmethyl)-1H-pyrazol-3-yl]phenoxy}methyl)quinoline, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Pandit, J, Marr, E.S. | | Deposit date: | 2009-06-08 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a Novel Class of Phosphodiesterase 10A Inhibitors and Identification of Clinical Candidate 2-[4-(1-Methyl-4-pyridin-4-yl-1H-pyrazol-3-yl)-phenoxymethyl]-quinoline (PF-2545920) for the Treatment of Schizophrenia
J.Med.Chem., 52, 2009
|
|
3HSZ
 
 | | Crystal structure of E. coli HPPK(F123A) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-06-11 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Pterin-binding site mutation Y53A, N55A or F123A and activity of E. coli HPPK
To be Published
|
|
3HTH
 
 | |
1P6A
 
 | | STRUCTURAL BASIS FOR VARIATION IN ADENOVIRUS AFFINITY FOR THE CELLULAR RECEPTOR CAR (S489Y MUTANT) | | Descriptor: | Coxsackievirus and adenovirus receptor, Fiber protein | | Authors: | Howitt, J, Bewley, M.C, Graziano, V, Flanagan, J.M, Freimuth, P. | | Deposit date: | 2003-04-29 | | Release date: | 2004-05-11 | | Last modified: | 2018-08-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for variation in adenovirus affinity for the cellular coxsackievirus and adenovirus receptor.
J.Biol.Chem., 278, 2003
|
|
3HVL
 
 | | Tethered PXR-LBD/SRC-1p complexed with SR-12813 | | Descriptor: | Pregnane X receptor, Linker, Steroid receptor coactivator 1, ... | | Authors: | Lesburg, C.A, Wang, W, Prosise, W.W, Chen, J, Taremi, S.S, Le, H.V, Madison, V, Cui, X, Thomas, A, Cheng, K.C. | | Deposit date: | 2009-06-16 | | Release date: | 2009-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Construction and characterization of a fully active PXR/SRC-1 tethered protein with increased stability
Protein Eng.Des.Sel., 21, 2008
|
|
3IC5
 
 | |
3IAK
 
 | | Crystal structure of human phosphodiesterase 4d (PDE4d) with papaverine. | | Descriptor: | 1-(3,4-DIMETHOXYBENZYL)-6,7-DIMETHOXYISOQUINOLINE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Cheng, R.K.Y, Crawley, L, Barker, J, Wood, M, Felicetti, B, Whittaker, M. | | Deposit date: | 2009-07-14 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human phosphodiesterase 4d (PDE4d) with papaverine.
To be Published
|
|
3IHJ
 
 | | Human alanine aminotransferase 2 in complex with PLP | | Descriptor: | Alanine aminotransferase 2, PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Wisniewska, M, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Kotzsch, A, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Schutz, P, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-30 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human glutamate pyruvate transaminase 2
To be Published
|
|
