8HTE
 
 | | Crystal structure of an effector mutant in complex with ubiquitin | | 分子名称: | GLYCEROL, NAD(+)--protein-threonine ADP-ribosyltransferase, NICOTINAMIDE, ... | | 著者 | Tan, J, Wang, X, Zhou, Y, Zhu, Y. | | 登録日 | 2022-12-21 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.307 Å) | | 主引用文献 | Molecular basis of threonine ADP-ribosylation of ubiquitin by bacterial ARTs.
Nat.Chem.Biol., 20, 2024
|
|
1AFX
 
 | |
8HTF
 
 | | Crystal structure of an effector in complex with ubiquitin | | 分子名称: | NAD(+)--protein-threonine ADP-ribosyltransferase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Ubiquitin-40S ribosomal protein S27a (Fragment) | | 著者 | Tan, J, Wang, X, Zhou, Y, Zhu, Y. | | 登録日 | 2022-12-21 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.151 Å) | | 主引用文献 | Molecular basis of threonine ADP-ribosylation of ubiquitin by bacterial ARTs.
Nat.Chem.Biol., 20, 2024
|
|
8HM5
 
 | |
1ABV
 
 | | N-TERMINAL DOMAIN OF THE DELTA SUBUNIT OF THE F1F0-ATP SYNTHASE FROM ESCHERICHIA COLI, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | DELTA SUBUNIT OF THE F1F0-ATP SYNTHASE | | 著者 | Wilkens, S, Dunn, S.D, Chandler, J, Dahlquist, F.W, Capaldi, R.A. | | 登録日 | 1997-01-29 | | 公開日 | 1997-07-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the N-terminal domain of the delta subunit of the E. coli ATPsynthase.
Nat.Struct.Biol., 4, 1997
|
|
4P86
 
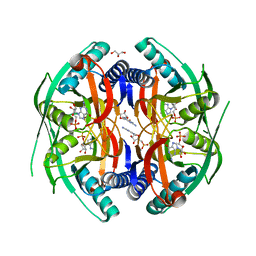 | | Structure of PyrR protein from Bacillus subtilis with GMP | | 分子名称: | Bifunctional protein PyrR, GLYCEROL, GUANOSINE-5'-MONOPHOSPHATE | | 著者 | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | 登録日 | 2014-03-30 | | 公開日 | 2014-12-17 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
4P8P
 
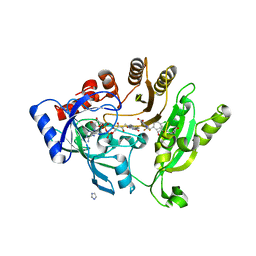 | | Crystal structure of M. tuberculosis DprE1 in complex with the non-covalent inhibitor QN127 | | 分子名称: | (2R)-2-{[(2R)-2-{[(2R)-2-hydroxypropyl]oxy}propyl]oxy}propan-1-ol, 3-[(4-chlorobenzyl)amino]-6-(trifluoromethyl)quinoxaline-2-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Neres, J, Pojer, F, Cole, S.T. | | 登録日 | 2014-03-31 | | 公開日 | 2014-12-10 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | 2-Carboxyquinoxalines Kill Mycobacterium tuberculosis through Noncovalent Inhibition of DprE1.
Acs Chem.Biol., 10, 2015
|
|
5E8C
 
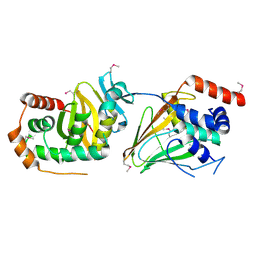 | | pseudorabies virus nuclear egress complex, pUL31, pUL34 | | 分子名称: | CHLORIDE ION, UL31, UL34 protein, ... | | 著者 | Zeev-Ben-Mordehai, T, Cheleski, J, Whittle, C, El Omari, K, Harlos, K, Hagen, C, Klupp, B, Mettenleiter, T.C, Gruenewald, K. | | 登録日 | 2015-10-14 | | 公開日 | 2015-12-23 | | 最終更新日 | 2016-01-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal Structure of the Herpesvirus Nuclear Egress Complex Provides Insights into Inner Nuclear Membrane Remodeling.
Cell Rep, 13, 2015
|
|
5E8L
 
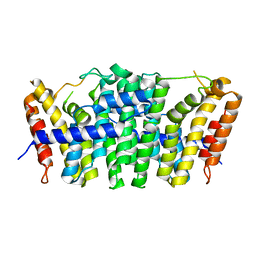 | | Crystal structure of geranylgeranyl pyrophosphate synthase 11 from Arabidopsis thaliana | | 分子名称: | Heterodimeric geranylgeranyl pyrophosphate synthase large subunit 1, chloroplastic | | 著者 | Wang, C, Chen, Q, Fan, D, Li, J, Wang, G, Zhang, P. | | 登録日 | 2015-10-14 | | 公開日 | 2015-11-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.807 Å) | | 主引用文献 | Structural Analyses of Short-Chain Prenyltransferases Identify an Evolutionarily Conserved GFPPS Clade in Brassicaceae Plants.
Mol Plant, 9, 2016
|
|
1A57
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF A HELIX-LESS VARIANT OF INTESTINAL FATTY ACID BINDING PROTEIN, NMR, 20 STRUCTURES | | 分子名称: | INTESTINAL FATTY ACID-BINDING PROTEIN | | 著者 | Steele, R.A, Emmert, D.A, Kao, J, Hodsdon, M.E, Frieden, C, Cistola, D.P. | | 登録日 | 1998-02-20 | | 公開日 | 1998-05-27 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The three-dimensional structure of a helix-less variant of intestinal fatty acid-binding protein.
Protein Sci., 7, 1998
|
|
1ACD
 
 | | V32D/F57H MUTANT OF MURINE ADIPOCYTE LIPID BINDING PROTEIN | | 分子名称: | ADIPOCYTE LIPID BINDING PROTEIN | | 著者 | Ory, J, Kane, C.D, Simpson, M, Banaszak, L.J, Bernlohr, D.A. | | 登録日 | 1997-02-06 | | 公開日 | 1997-06-16 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Biochemical and crystallographic analyses of a portal mutant of the adipocyte lipid-binding protein.
J.Biol.Chem., 272, 1997
|
|
4PA5
 
 | | Tgl - a bacterial spore coat transglutaminase - cystamine complex | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-ETHANETHIOL, CITRATE ANION, ... | | 著者 | Brito, J.A, Placido, D, Fernandes, C.G, Lousa, D, Isidro, A, Soares, C.M, Pohl, J, Carrondo, M.A, Henriques, A.O, Archer, M. | | 登録日 | 2014-04-07 | | 公開日 | 2015-09-30 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Structural and Functional Characterization of an Ancient Bacterial Transglutaminase Sheds Light on the Minimal Requirements for Protein Cross-Linking.
Biochemistry, 54, 2015
|
|
5E4Y
 
 | | Orthorhombic structure of the acetyl esterase MekB | | 分子名称: | Homoserine O-acetyltransferase | | 著者 | Niefind, K, Toelzer, C, Pal, S, Watzlawick, H, Altenbuchner, J. | | 登録日 | 2015-10-07 | | 公開日 | 2015-12-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A novel esterase subfamily with alpha / beta-hydrolase fold suggested by structures of two bacterial enzymes homologous to l-homoserine O-acetyl transferases.
Febs Lett., 590, 2016
|
|
4PAU
 
 | | Hypothetical protein SA1058 from S. aureus. | | 分子名称: | CHLORIDE ION, Nitrogen regulatory protein A | | 著者 | Battaile, K.P, Wu-Brown, J, Romanov, V, Jones, K, Lam, R, Pai, E.F, Chirgadze, N.Y. | | 登録日 | 2014-04-10 | | 公開日 | 2015-05-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Hypothetical protein SA1058 from S. aureus.
To Be Published
|
|
5E66
 
 | | The complex structure of Hemagglutinin-esterase-fusion mutant protein from the influenza D virus with receptor analog 9-N-Ac-Sia | | 分子名称: | (6R)-5-acetamido-6-[(1S,2S)-3-acetamido-1,2-dihydroxypropyl]-3,5-dideoxy-beta-L-threo-hex-2-ulopyranosonic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Song, H, Qi, J, Shi, Y, Gao, G.F. | | 登録日 | 2015-10-09 | | 公開日 | 2016-03-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | An Open Receptor-Binding Cavity of Hemagglutinin-Esterase-Fusion Glycoprotein from Newly-Identified Influenza D Virus: Basis for Its Broad Cell Tropism
PLoS Pathog., 12, 2016
|
|
4PBW
 
 | | Crystal structure of chicken receptor protein tyrosine phosphatase sigma in complex with TrkC | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NT-3 growth factor receptor, Protein-tyrosine phosphatase CRYPalpha1 isoform | | 著者 | Coles, C.H, Mitakidis, N, Zhang, P, Elegheert, J, Lu, W, Stoker, A.W, Nakagawa, T, Craig, A.M, Jones, E.Y, Aricescu, A.R. | | 登録日 | 2014-04-14 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Structural basis for extracellular cis and trans RPTP sigma signal competition in synaptogenesis.
Nat Commun, 5, 2014
|
|
4PCE
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with compound B13 | | 分子名称: | 1,2-ETHANEDIOL, 1-benzyl-2-ethyl-1,5,6,7-tetrahydro-4H-indol-4-one, Bromodomain-containing protein 4 | | 著者 | Dong, J, Caflisch, A. | | 登録日 | 2014-04-15 | | 公開日 | 2014-05-07 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.293 Å) | | 主引用文献 | Discovery of BRD4 bromodomain inhibitors by fragment-based high-throughput docking.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
1AFH
 
 | | LIPID TRANSFER PROTEIN FROM MAIZE SEEDLINGS, NMR, 15 STRUCTURES | | 分子名称: | MAIZE NONSPECIFIC LIPID TRANSFER PROTEIN | | 著者 | Gomar, J, Petit, M.C, Sodano, P, Sy, D, Marion, D, Kader, J.C, Vovelle, F, Ptak, M. | | 登録日 | 1997-03-07 | | 公開日 | 1997-05-15 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and lipid binding of a nonspecific lipid transfer protein extracted from maize seeds.
Protein Sci., 5, 1996
|
|
1A9M
 
 | | G48H MUTANT OF HIV-1 PROTEASE IN COMPLEX WITH A PEPTIDIC INHIBITOR U-89360E | | 分子名称: | HIV-1 PROTEASE, N-[[1-[N-ACETAMIDYL]-[1-CYCLOHEXYLMETHYL-2-HYDROXY-4-ISOPROPYL]-BUT-4-YL]-CARBONYL]-GLUTAMINYL-ARGINYL-AMIDE | | 著者 | Hong, L, Zhang, X.-J, Foundling, S, Hartsuck, J.A, Tang, J. | | 登録日 | 1998-04-08 | | 公開日 | 1998-06-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of a G48H mutant of HIV-1 protease explains how glycine-48 replacements produce mutants resistant to inhibitor drugs.
FEBS Lett., 420, 1997
|
|
1WYB
 
 | | Structure of 6-aminohexanoate-dimer hydrolase | | 分子名称: | 6-aminohexanoate-dimer hydrolase | | 著者 | Negoro, S, Ohki, T, Shibata, N, Mizuno, N, Wakitani, Y, Tsurukame, J, Matsumoto, K, Kawamoto, I, Takeo, M, Higuchi, Y. | | 登録日 | 2005-02-09 | | 公開日 | 2005-09-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of 6-aminohexanoate-dimer hydrolase
To be Published
|
|
5DUM
 
 | | Crystal structure of influenza A virus H5 hemagglutinin globular head in complex with the Fab of antibody 65C6 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 65C6 Heavy Chain, 65C6 Light Chain, ... | | 著者 | Sun, J, Zuo, T, Wang, G, Zhou, P, Zhang, L, Wang, X. | | 登録日 | 2015-09-19 | | 公開日 | 2015-12-16 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.003 Å) | | 主引用文献 | Comprehensive analysis of antibody recognition in convalescent humans from highly pathogenic avian influenza H5N1 infection
Nat Commun, 6, 2015
|
|
5DUQ
 
 | | Active human c1-inhibitor in complex with dextran sulfate | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Plasma protease C1 inhibitor, SULFITE ION, ... | | 著者 | Dijk, M, Holkers, J, Voskamp, P, Giannetti, B.M, Waterreus, W.J, van Veen, H.A, Pannu, N.S. | | 登録日 | 2015-09-20 | | 公開日 | 2016-08-31 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | How Dextran Sulfate Affects C1-inhibitor Activity: A Model for Polysaccharide Potentiation.
Structure, 24, 2016
|
|
4PQV
 
 | | Crystal structure of an Xrn1-resistant RNA from the 3' untranslated region of a flavivirus (Murray Valley Encephalitis virus) | | 分子名称: | MAGNESIUM ION, XRN1-resistant flaviviral RNA | | 著者 | Chapman, E.G, Costantino, D.A, Rabe, J.L, Moon, S.L, Wilusz, J, Nix, J.C, Kieft, J.S. | | 登録日 | 2014-03-04 | | 公開日 | 2014-04-30 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.463 Å) | | 主引用文献 | The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production.
Science, 344, 2014
|
|
1AMC
 
 | |
3N9T
 
 | | Cryatal structure of Hydroxyquinol 1,2-dioxygenase from Pseudomonas putida DLL-E4 | | 分子名称: | 1-HEPTADECANOYL-2-TRIDECANOYL-3-GLYCEROL-PHOSPHONYL CHOLINE, CITRATE ANION, FE (III) ION, ... | | 著者 | Liu, W, Shen, W, Fang, P, Li, J, Cui, Z. | | 登録日 | 2010-05-31 | | 公開日 | 2010-08-04 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Cryatal structure of Hydroxyquinol 1,2-dioxygenase from Pseudomonas putida DLL-E4
To be Published
|
|
