1GF9
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | 登録日 | 2000-12-04 | | 公開日 | 2000-12-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1GFJ
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | 登録日 | 2000-12-04 | | 公開日 | 2000-12-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
7ZRA
 
 | | Crystal structure of E.coli LexA in complex with nanobody NbSOS1(Nb14497) | | 分子名称: | 1,2-ETHANEDIOL, LexA repressor, Nanobody NbSOS1 (Nb14497) | | 著者 | Maso, L, Vascon, F, Chinellato, M, Pardon, E, Steyaert, J, Angelini, A, Tondi, D, Cendron, L. | | 登録日 | 2022-05-04 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Nanobodies targeting LexA autocleavage disclose a novel suppression strategy of SOS-response pathway.
Structure, 30, 2022
|
|
8KG9
 
 | | Yeast replisome in state III | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, DNA (61-mer), ... | | 著者 | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | 登録日 | 2023-08-17 | | 公開日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (4.52 Å) | | 主引用文献 | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
5JAG
 
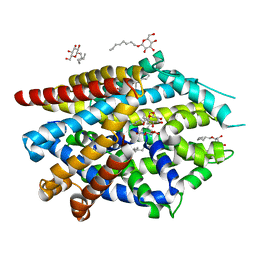 | | LeuT T354H mutant in the outward-oriented, Na+-free Return State | | 分子名称: | Transporter, octyl beta-D-glucopyranoside | | 著者 | Malinauskaite, L, Sahin, C, Said, S, Grouleff, J, Shahsavar, A, Bjerregaard, H, Noer, P, Severinsen, K, Boesen, T, Schiott, B, Sinning, S, Nissen, P. | | 登録日 | 2016-04-12 | | 公開日 | 2016-06-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | A conserved leucine occupies the empty substrate site of LeuT in the Na(+)-free return state.
Nat Commun, 7, 2016
|
|
7K3M
 
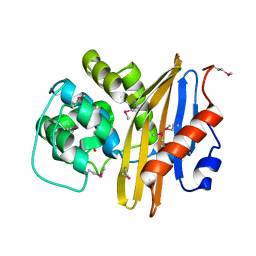 | | Crystal Structure of the Beta Lactamase Class D from Chitinophaga pinensis by Serial Crystallography | | 分子名称: | Beta-lactamase | | 著者 | Kim, Y, Sherrell, D.A, Johnson, J, Lavens, A, Maltseva, N, Endres, M, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-09-11 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of the Beta Lactamase Class D from Chitinophaga pinensis by Serial Crystallography
To Be Published
|
|
8A8U
 
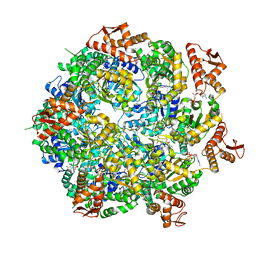 | | Mycobacterium tuberculosis ClpC1 hexamer structure | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpC1, Bound polypeptide | | 著者 | Felix, J, Fraga, H, Gragera, M, Bueno, T, Weinhaeupl, K. | | 登録日 | 2022-06-24 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.62 Å) | | 主引用文献 | Structure of the drug target ClpC1 unfoldase in action provides insights on antibiotic mechanism of action.
J.Biol.Chem., 298, 2022
|
|
8A8V
 
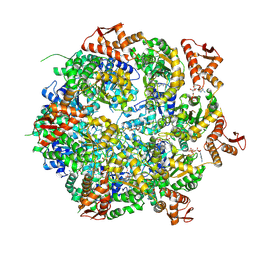 | | Mycobacterium tuberculosis ClpC1 hexamer structure bound to the natural product antibiotic Cyclomarin | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpC1, Bound polypeptide | | 著者 | Felix, J, Fraga, H, Gragera, M, Bueno, T, Weinhaeupl, K. | | 登録日 | 2022-06-24 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.34 Å) | | 主引用文献 | Structure of the drug target ClpC1 unfoldase in action provides insights on antibiotic mechanism of action.
J.Biol.Chem., 298, 2022
|
|
7JOY
 
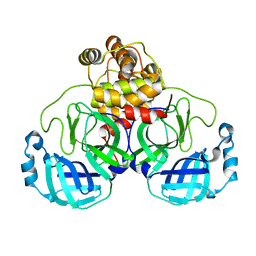 | |
8A8W
 
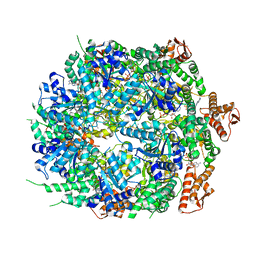 | | Mycobacterium tuberculosis ClpC1 hexamer structure bound to the natural product antibiotic Ecumycin (class 1) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpC1, Bound polypeptide | | 著者 | Felix, J, Fraga, H, Gragera, M, Bueno, T, Weinhaeupl, K. | | 登録日 | 2022-06-24 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (4.29 Å) | | 主引用文献 | Structure of the drug target ClpC1 unfoldase in action provides insights on antibiotic mechanism of action.
J.Biol.Chem., 298, 2022
|
|
5DAB
 
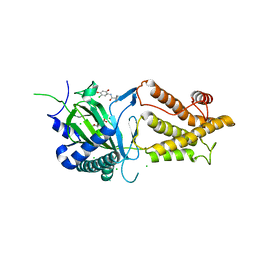 | | Crystal structure of FTO-IN115 | | 分子名称: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase FTO, CHLORIDE ION, ... | | 著者 | Chai, J, Zhou, B, Liu, W, Han, Z, Niu, T. | | 登録日 | 2015-08-19 | | 公開日 | 2016-08-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of FTO-IN115
To Be Published
|
|
1K4B
 
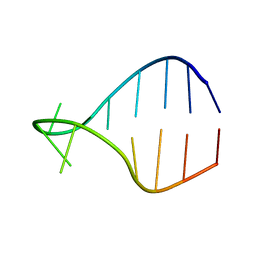 | | STRUCTURE OF AGUU RNA TETRALOOP, NMR, 20 STRUCTURES | | 分子名称: | 5'-R(*GP*GP*UP*UP*CP*AP*GP*UP*UP*GP*AP*AP*CP*C)-3' | | 著者 | Wu, H, Yang, P.K, Butcher, S.E, Kang, S, Chanfreau, G, Feigon, J. | | 登録日 | 2001-10-07 | | 公開日 | 2001-12-19 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A novel family of RNA tetraloop structure forms the recognition site for Saccharomyces cerevisiae RNase III.
EMBO J., 20, 2001
|
|
8KG6
 
 | | Yeast replisome in state I | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, Chromosome segregation in meiosis protein 3, ... | | 著者 | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | 登録日 | 2023-08-17 | | 公開日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
5JBW
 
 | | Crystal structure of LiuC | | 分子名称: | 3-hydroxybutyryl-CoA dehydratase | | 著者 | Bock, T, Reichelt, J, Mueller, R, Blankenfeldt, W. | | 登録日 | 2016-04-14 | | 公開日 | 2016-06-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | The Structure of LiuC, a 3-Hydroxy-3-Methylglutaconyl CoA Dehydratase Involved in Isovaleryl-CoA Biosynthesis in Myxococcus xanthus, Reveals Insights into Specificity and Catalysis.
Chembiochem, 17, 2016
|
|
1K9S
 
 | | PURINE NUCLEOSIDE PHOSPHORYLASE FROM E. COLI IN COMPLEX WITH FORMYCIN A DERIVATIVE AND PHOSPHATE | | 分子名称: | 2-(7-AMINO-6-METHYL-3H-PYRAZOLO[4,3-D]PYRIMIDIN-3-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, 2-HYDROXYMETHYL-5-(7-METHYLAMINO-3H-PYRAZOLO[4,3-D]PYRIMIDIN-3-YL)-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, ... | | 著者 | Koellner, G, Bzowska, A, Wielgus-Kutrowska, B, Luic, M, Steiner, T, Saenger, W, Stepinski, J. | | 登録日 | 2001-10-30 | | 公開日 | 2001-11-28 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Open and closed conformation of the E. coli purine nucleoside phosphorylase active center and implications for the catalytic mechanism.
J.Mol.Biol., 315, 2002
|
|
3C7V
 
 | |
7LNJ
 
 | |
5JJR
 
 | | Dengue 3 NS5 protein with compound 29 | | 分子名称: | 1,2-ETHANEDIOL, 5-[5-(3-hydroxyprop-1-yn-1-yl)thiophen-2-yl]-4-methoxy-2-methyl-N-[(quinolin-8-yl)sulfonyl]benzamide, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Lescar, J, El Sahili, A. | | 登録日 | 2016-04-25 | | 公開日 | 2016-07-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Potent Allosteric Dengue Virus NS5 Polymerase Inhibitors: Mechanism of Action and Resistance Profiling
Plos Pathog., 12, 2016
|
|
1O8Q
 
 | | The active site of the molybdenum cofactor biosenthetic protein domain Cnx1G | | 分子名称: | MOLYBDOPTERIN BIOSYNTHESIS CNX1 PROTEIN | | 著者 | Kuper, J, Winking, J, Hecht, H.J, Schwarz, G, Mendel, R.R. | | 登録日 | 2002-11-28 | | 公開日 | 2003-02-27 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The Active Site of the Molybdenum Cofactor Biosynthetic Protein Domain Cnx1G
Arch.Biochem.Biophys., 411, 2003
|
|
4R11
 
 | | A conserved phosphorylation switch controls the interaction between cadherin and beta-catenin in vitro and in vivo | | 分子名称: | Cadherin-related hmr-1, IODIDE ION, Protein humpback-2 | | 著者 | Choi, H.-J, Loveless, T, Lynch, A, Bang, I, Hardin, J, Weis, W.I. | | 登録日 | 2014-08-03 | | 公開日 | 2015-04-29 | | 実験手法 | X-RAY DIFFRACTION (2.789 Å) | | 主引用文献 | A Conserved Phosphorylation Switch Controls the Interaction between Cadherin and beta-Catenin In Vitro and In Vivo
Dev.Cell, 33, 2015
|
|
5JCS
 
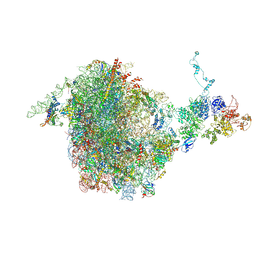 | | CRYO-EM STRUCTURE OF THE RIX1-REA1 PRE-60S PARTICLE | | 分子名称: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | 著者 | Barrio-Garcia, C, Thoms, M, Flemming, D, Kater, L, Berninghausen, O, Bassler, J, Beckmann, R, Hurt, E. | | 登録日 | 2016-04-15 | | 公開日 | 2016-11-16 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (9.5 Å) | | 主引用文献 | Architecture of the Rix1-Rea1 checkpoint machinery during pre-60S-ribosome remodeling
Nat.Struct.Mol.Biol., 23, 2016
|
|
1FTS
 
 | |
3C9P
 
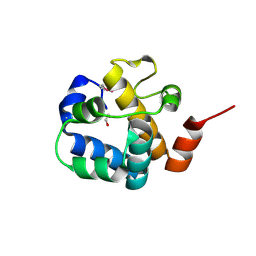 | | Crystal structure of uncharacterized protein SP1917 | | 分子名称: | 1,2-ETHANEDIOL, Uncharacterized protein SP1917 | | 著者 | Chang, C, Zhou, M, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-02-18 | | 公開日 | 2008-02-26 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Crystal structure of uncharacterized protein SP1917.
To be Published
|
|
1K73
 
 | | Co-crystal Structure of Anisomycin Bound to the 50S Ribosomal Subunit | | 分子名称: | 23S RRNA, 5S RRNA, ANISOMYCIN, ... | | 著者 | Hansen, J, Ban, N, Nissen, P, Moore, P.B, Steitz, T.A. | | 登録日 | 2001-10-18 | | 公開日 | 2003-07-22 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | Structures of Five Antibiotics Bound at the Peptidyl Transferase Center of
the Large Ribosomal Subunit
J.Mol.Biol., 330, 2003
|
|
3BOG
 
 | | Snow Flea Antifreeze Protein Quasi-racemate | | 分子名称: | 6.5 kDa glycine-rich antifreeze protein, UNKNOWN LIGAND | | 著者 | Pentelute, B.L, Kent, S.B.H, Gates, Z.P, Tereshko, V, Kossiakoff, A.A, Kurutz, J, Dashnau, J, Vaderkooi, J.M. | | 登録日 | 2007-12-17 | | 公開日 | 2008-09-23 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | X-ray structure of snow flea antifreeze protein determined by racemic crystallization of synthetic protein enantiomers
J.Am.Chem.Soc., 130, 2008
|
|
