4WFJ
 
 | | Crystal structure of PET-degrading cutinase Cut190 S226P mutant in Ca(2+)-bound state at 1.75 angstrom resolution | | 分子名称: | CALCIUM ION, CHLORIDE ION, Cutinase | | 著者 | Miyakawa, T, Mizushima, H, Ohtsuka, J, Oda, M, Kawai, F, Tanokura, M. | | 登録日 | 2014-09-15 | | 公開日 | 2014-12-24 | | 最終更新日 | 2020-01-29 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural basis for the Ca(2+)-enhanced thermostability and activity of PET-degrading cutinase-like enzyme from Saccharomonospora viridis AHK190.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4WFK
 
 | | Crystal structure of PET-degrading cutinase Cut190 S226P mutant in Ca(2+)-bound state at 2.35 angstrom resolution | | 分子名称: | CALCIUM ION, CHLORIDE ION, Cutinase | | 著者 | Miyakawa, T, Mizushima, H, Ohtsuka, J, Oda, M, Kawai, F, Tanokura, M. | | 登録日 | 2014-09-15 | | 公開日 | 2014-12-24 | | 最終更新日 | 2020-01-29 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural basis for the Ca(2+)-enhanced thermostability and activity of PET-degrading cutinase-like enzyme from Saccharomonospora viridis AHK190.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4WK6
 
 | | Crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase (FabG) (G141A) from Vibrio cholerae in complex with NADPH | | 分子名称: | 3-oxoacyl-[acyl-carrier protein] reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PENTAETHYLENE GLYCOL | | 著者 | Hou, J, Zheng, H, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-10-01 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
4WM7
 
 | | Crystal Structure of Human Enterovirus D68 in Complex with Pleconaril | | 分子名称: | 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-TRIFLUOROMETHYL-[1,2,4]OXADIAZOLE, VP1, VP2, ... | | 著者 | Liu, Y, Sheng, J, Fokine, A, Meng, G, Long, F, Kuhn, R.J, Rossmann, M.G. | | 登録日 | 2014-10-08 | | 公開日 | 2015-01-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Virus structure. Structure and inhibition of EV-D68, a virus that causes respiratory illness in children.
Science, 347, 2015
|
|
6NMU
 
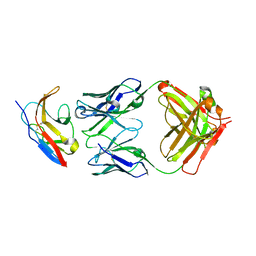 | | Kick-Off Fab 115 anti-SIRP-alpha antibody in complex with SIRP-alpha Variant 1 | | 分子名称: | Fab 115 anti-SIRP-alpha antibody Variable Heavy Chain, Fab 115 anti-SIRP-alpha antibody Variable Light Chain, Tyrosine-protein phosphatase non-receptor type substrate 1 | | 著者 | Wibowo, A.S, Carter, J.J, Sim, J. | | 登録日 | 2019-01-11 | | 公開日 | 2019-08-07 | | 最終更新日 | 2019-08-14 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Discovery of high affinity, pan-allelic, and pan-mammalian reactive antibodies against the myeloid checkpoint receptor SIRP alpha.
Mabs, 11, 2019
|
|
4WP0
 
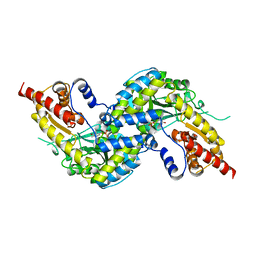 | | Crystal structure of human kynurenine aminotransferase-I with a C-terminal V5-hexahistidine tag | | 分子名称: | Kynurenine--oxoglutarate transaminase 1 | | 著者 | Nadvi, N.A, Salam, N.K, Park, J, Akladios, F.N, Kapoor, V, Collyer, C.A, Gorrell, M.D, Church, W.B. | | 登録日 | 2014-10-17 | | 公開日 | 2014-11-19 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of human kynurenine aminotransferase-I in a novel space group
To be published
|
|
6NQB
 
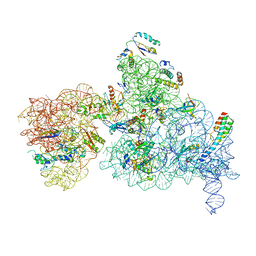 | |
8DGB
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Q192T Mutant in Complex with Inhibitor GC376 | | 分子名称: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Jacobs, L.M.C, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-23 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.87 Å) | | 主引用文献 | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
4WTR
 
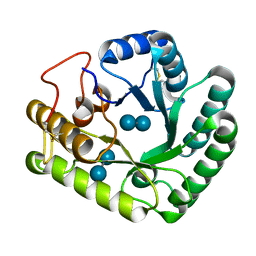 | | Active-site mutant of Rhizomucor miehei beta-1,3-glucanosyltransferase in complex with laminaribiose | | 分子名称: | beta-1,3-glucanosyltransferase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | 著者 | Qin, Z, Yan, Q, Lei, J, Yang, S, Jiang, Z. | | 登録日 | 2014-10-30 | | 公開日 | 2015-08-12 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | The first crystal structure of a glycoside hydrolase family 17 beta-1,3-glucanosyltransferase displays a unique catalytic cleft.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WEV
 
 | | Crystal structure of human AKR1B10 complexed with NADP+ and sulindac | | 分子名称: | Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [(1Z)-5-fluoro-2-methyl-1-{4-[methylsulfinyl]benzylidene}-1H-inden-3-yl]acetic acid | | 著者 | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Crespo, I, Porte, S, Pares, X, Farres, J, Podjarny, A. | | 登録日 | 2014-09-11 | | 公開日 | 2015-01-14 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.453 Å) | | 主引用文献 | Structural analysis of sulindac as an inhibitor of aldose reductase and AKR1B10.
Chem.Biol.Interact., 234, 2015
|
|
8DCZ
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) M165Y Mutant in Complex with Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-17 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DD1
 
 | | SARS-CoV-2 Main Protease (Mpro) H164N Mutant in Complex with Inhibitor GC376 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Butler, S.G, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-17 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DFN
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) H164N Mutant | | 分子名称: | 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Butler, S.G, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-22 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DFE
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144L Mutant | | 分子名称: | 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Jacobs, L.M.C, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-22 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DD9
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144L Mutant in Complex with Inhibitor GC376 | | 分子名称: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Jacobs, L.M.C, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-17 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
4WJN
 
 | | Crystal structure of SUMO1 in complex with phosphorylated PML | | 分子名称: | Protein PML, Small ubiquitin-related modifier 1 | | 著者 | Cappadocia, L, Mascle, X.H, Bourdeau, V, Tremblay-Belzile, S, Chaker-Margot, M, Lussier-Price, M, Wada, J, Sakaguchi, K, Aubry, M, Ferbeyre, G, Omichinski, J.G. | | 登録日 | 2014-10-01 | | 公開日 | 2014-12-31 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural and Functional Characterization of the Phosphorylation-Dependent Interaction between PML and SUMO1.
Structure, 23, 2015
|
|
4WJP
 
 | | Crystal Structure of SUMO1 in complex with phosphorylated Daxx | | 分子名称: | Daxx, Small ubiquitin-related modifier 1 | | 著者 | Cappadocia, L, Mascle, X.H, Bourdeau, V, Tremblay-Belzile, S, Chaker-Margot, M, Lussier-Price, M, Wada, J, Sakaguchi, K, Aubry, M, Ferbeyre, G, Omichinski, J.G. | | 登録日 | 2014-10-01 | | 公開日 | 2014-12-31 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and Functional Characterization of the Phosphorylation-Dependent Interaction between PML and SUMO1.
Structure, 23, 2015
|
|
4WSR
 
 | | The crystal structure of hemagglutinin form A/chicken/New York/14677-13/1998 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | 著者 | Yang, H, Carney, P.J, Chang, J.C, Villanueva, J.M, Stevens, J. | | 登録日 | 2014-10-28 | | 公開日 | 2015-02-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure and receptor binding preferences of recombinant hemagglutinins from avian and human h6 and h10 influenza a virus subtypes.
J.Virol., 89, 2015
|
|
4WSU
 
 | | The crystal structure of hemagglutinin from A/Taiwan/1/2013 in complex with 3'SLN | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain, ... | | 著者 | Yang, H, Carney, P.J, Chang, J.C, Villanueva, J.M, Stevens, J. | | 登録日 | 2014-10-28 | | 公開日 | 2015-02-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure and receptor binding preferences of recombinant hemagglutinins from avian and human h6 and h10 influenza a virus subtypes.
J.Virol., 89, 2015
|
|
4WSX
 
 | | The crystal structure of hemagglutinin from A/Jiangxi-Donghu/346/2013 influenza virus | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | 著者 | Yang, H, Carney, P.J, Chang, J.C, Villanueva, J.M, Stevens, J. | | 登録日 | 2014-10-28 | | 公開日 | 2015-02-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure and receptor binding preferences of recombinant hemagglutinins from avian and human h6 and h10 influenza a virus subtypes.
J.Virol., 89, 2015
|
|
4WPO
 
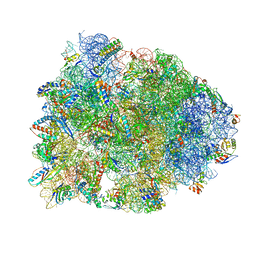 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with elongation factor G in the pre-translocational state | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Lin, J, Gagnon, M.G, Steitz, T.A. | | 登録日 | 2014-10-20 | | 公開日 | 2015-01-28 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Conformational Changes of Elongation Factor G on the Ribosome during tRNA Translocation.
Cell, 160, 2015
|
|
4X6E
 
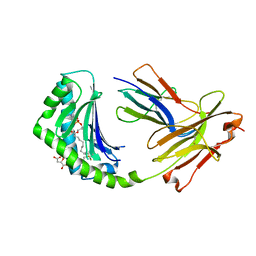 | | CD1a binary complex with lysophosphatidylcholine | | 分子名称: | (4R,7R,18Z)-4,7-dihydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphaheptacos-18-en-1-aminium 4-oxide, Beta-2-microglobulin, D-MALATE, ... | | 著者 | Birkinshaw, R.W, Rossjohn, J. | | 登録日 | 2014-12-08 | | 公開日 | 2015-01-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | alpha beta T cell antigen receptor recognition of CD1a presenting self lipid ligands.
Nat.Immunol., 16, 2015
|
|
4X6H
 
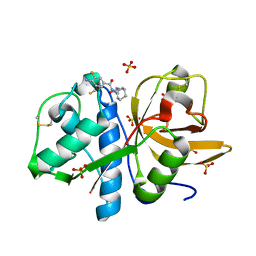 | | Development of N-(Functionalized benzoyl)-homocycloleucyl-glycinonitriles as Potent Cathepsin K Inhibitors. | | 分子名称: | 4-amino-3-fluoro-N-(1-{[(2Z)-2-iminoethyl]carbamoyl}cyclohexyl)benzamide, 4-amino-N-{1-[(cyanomethyl)carbamoyl]cyclohexyl}-3-fluorobenzamide, Cathepsin K, ... | | 著者 | Borisek, J, Mohar, B, Vizovisek, M, Sosnowski, P, Turk, D, Turk, B, Novic, M. | | 登録日 | 2014-12-08 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Development of N-(Functionalized benzoyl)-homocycloleucyl-glycinonitriles as Potent Cathepsin K Inhibitors.
J.Med.Chem., 58, 2015
|
|
4WTP
 
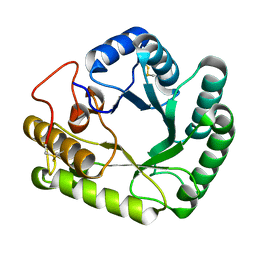 | | Crystal structure of glycoside hydrolase family 17 beta-1,3-glucanosyltransferase from Rhizomucor miehei | | 分子名称: | 1,2-ETHANEDIOL, beta-1,3-glucanosyltransferase | | 著者 | Qin, Z, Yan, Q, Lei, J, Yang, S, Jiang, Z. | | 登録日 | 2014-10-30 | | 公開日 | 2015-08-12 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | The first crystal structure of a glycoside hydrolase family 17 beta-1,3-glucanosyltransferase displays a unique catalytic cleft.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WTS
 
 | | Active-site mutant of Rhizomucor miehei beta-1,3-glucanosyltransferase in complex with laminaritriose | | 分子名称: | beta-1,3-glucanosyltransferase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | 著者 | Qin, Z, Yan, Q, Lei, J, Yang, S, Jiang, Z. | | 登録日 | 2014-10-30 | | 公開日 | 2015-08-12 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The first crystal structure of a glycoside hydrolase family 17 beta-1,3-glucanosyltransferase displays a unique catalytic cleft.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
