2ZOL
 
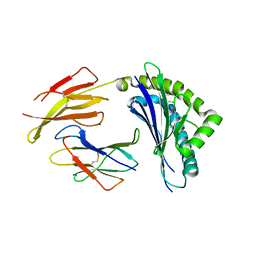 | | Crystal structure of H-2Db in complex with the W513S variant of JHMV epitope S510 | | Descriptor: | 9-meric peptide from Spike glycoprotein, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Theodossis, A, Dunstone, M.A, Rossjohn, J. | | Deposit date: | 2008-05-22 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and biological basis of CTL escape in coronavirus-infected mice.
J Immunol., 180, 2008
|
|
5IKY
 
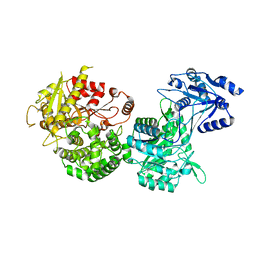 | |
5B4W
 
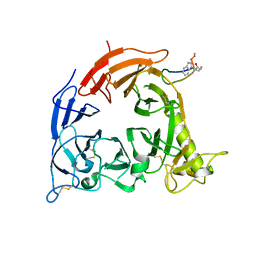 | | Crystal structure of Plexin inhibitor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-B1, Synthesized cyclic peptide | | Authors: | Matsunaga, Y, Kitago, Y, Arimori, T, Takagi, J. | | Deposit date: | 2016-04-19 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Allosteric Inhibition of a Semaphorin 4D Receptor Plexin B1 by a High-Affinity Macrocyclic Peptide
Cell Chem Biol, 23, 2016
|
|
8GZD
 
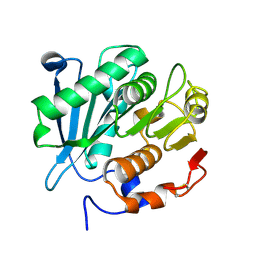 | |
2ZQV
 
 | |
2ZQU
 
 | |
7Z48
 
 | | Bottom part (C5) of bacteriophage SU10 capsid | | Descriptor: | Major head protein | | Authors: | Siborova, M, Fuzik, T, Prochazkova, M, Novacek, J, Plevka, P. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Tail proteins of phage SU10 reorganize into the nozzle for genome delivery.
Nat Commun, 13, 2022
|
|
5IOM
 
 | | Crystal Structure of Nucleoside Diphosphate Kinase from Schistosoma mansoni is space group P6322 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Torini, J.R.S, Romanello, L, Bird, L.E, Nettleship, J.E, Owens, R.J, Aller, P, DeMarco, R, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2016-03-08 | | Release date: | 2017-08-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of a Schistosoma mansoni NDPK expressed in sexual and digestive organs.
Mol.Biochem.Parasitol., 2019
|
|
5BMF
 
 | | Crystal Structure of a Theophylline binding antibody Fab fragment | | Descriptor: | 2-(5-{1-[1-(1,3-dimethyl-2,6-dioxo-2,3,6,7-tetrahydro-1H-purin-8-yl)-4,15-dioxo-8,11-dioxa-5,14-diazaicosan-20-yl]-3,3-dimethyl-6-sulfo-1,3-dihydro-2H-indol-2-ylidene}penta-1,3-dien-1-yl)-1-ethyl-3,3-dimethyl-3H-indolium-5-sulfonate, Fab fragment heavy chain, Fab fragment light chain | | Authors: | Bujotzek, A, Fuchs, A, Changtao, Q, Klostermann, S, Benz, J, Antes, I, Dengl, S, Hoffmann, E, Georges, G. | | Deposit date: | 2015-05-22 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | MoFvAb: Modeling the Fv region of antibodies.
Mabs, 7, 2015
|
|
5B60
 
 | | Crystal structure of PtLCIB4 S47R mutant, a homolog of the limiting CO2-inducible protein LCIB | | Descriptor: | CHLORIDE ION, PtLCIB4 S47R mutant, ZINC ION | | Authors: | Jin, S, Sun, J, Wunder, T, Tang, D, Mueller-Caja, O.M, Gao, Y. | | Deposit date: | 2016-05-24 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the LCIB protein family reveals a new group of beta-carbonic anhydrases
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5BNG
 
 | | monomer of TALE type homeobox transcription factor MEIS1 complexes with specific DNA | | Descriptor: | DNA (5'-D(P*AP*AP*TP*TP*AP*GP*CP*TP*GP*TP*CP*A)-3'), DNA (5'-D(P*TP*GP*AP*CP*AP*GP*CP*TP*AP*A)-3'), DNA (5'-D(P*TP*GP*AP*CP*AP*GP*CP*TP*AP*A-3'), ... | | Authors: | Morgunova, E, Jolma, A, Yin, Y, Nitta, K, Dave, K, Popov, A, Taipale, M, Enge, M, Kivioja, T, Taipale, J. | | Deposit date: | 2015-05-26 | | Release date: | 2015-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | DNA-dependent formation of transcription factor pairs alters their binding specificity.
Nature, 527, 2015
|
|
7Z44
 
 | | Portal of bacteriophage SU10 | | Descriptor: | Portal protein | | Authors: | Siborova, M, Fuzik, T, Prochazkova, M, Novacek, J, Plevka, P. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Tail proteins of phage SU10 reorganize into the nozzle for genome delivery.
Nat Commun, 13, 2022
|
|
5I7D
 
 | |
1SML
 
 | | METALLO BETA LACTAMASE L1 FROM STENOTROPHOMONAS MALTOPHILIA | | Descriptor: | PROTEIN (PENICILLINASE), ZINC ION | | Authors: | Ullah, J.H, Walsh, T.R, Taylor, I.A, Emery, D.C, Verma, C.S, Gamblin, S.J, Spencer, J. | | Deposit date: | 1998-09-22 | | Release date: | 1999-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of the L1 metallo-beta-lactamase from Stenotrophomonas maltophilia at 1.7 A resolution.
J.Mol.Biol., 284, 1998
|
|
5BOC
 
 | | Crystal structure of topoisomerase ParE inhibitor | | Descriptor: | 3-methyl-4-({3-[3-methyl-5-(trifluoromethyl)phenyl]-1H-pyrazol-5-yl}carbamoyl)benzoic acid, DNA topoisomerase 4 subunit B | | Authors: | Tan, Y.W, Chen, G.Y, Hung, A.W, Hill, J. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Application of Fragment-based Drug Discovery against DNA GyraseB
To be published
|
|
8HY5
 
 | | Structure of D-amino acid oxidase mutant R38H | | Descriptor: | 1,2-ETHANEDIOL, BENZOIC ACID, D-amino-acid oxidase, ... | | Authors: | Khan, S, Upadhyay, S, Dave, U, Kumar, A, Gomes, J. | | Deposit date: | 2023-01-05 | | Release date: | 2023-01-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mechanistic insights into ALS patient derived mutations in D-amino acid oxidase.
Int.J.Biol.Macromol., 256, 2023
|
|
5BOD
 
 | | Crystal structure of Streptococcus pneumonia ParE inhibitor | | Descriptor: | (2R)-N-[3-(3,5-dimethylphenyl)-1H-pyrazol-5-yl]-1,4-dioxane-2-carboxamide, DNA topoisomerase 4 subunit B | | Authors: | Tan, Y.W, Chen, G, Hung, A.W, Hill, J. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Application of Fragment-based Drug Discovery against DNA GyraseB
to be published
|
|
8I6L
 
 | | Crystal structure of horse spleen L-ferritin at -180deg Celsius. | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Maity, B, Tian, J, Abe, S, Ueno, T. | | Deposit date: | 2023-01-28 | | Release date: | 2023-03-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Atomic-level insights into a unique semi-clathrate hydrate formed in a confined environment of porous protein crystal.
Cryst.Growth Des., 2023
|
|
5IQ2
 
 | |
8I81
 
 | | Crystal structure of horse spleen L-ferritin A115G mutant at -180deg Celsius. | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Maity, B, Tian, J, Abe, S, Ueno, T. | | Deposit date: | 2023-02-02 | | Release date: | 2023-03-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Atomic-level insights into a unique semi-clathrate hydrate formed in a confined environment of porous protein crystal.
Cryst.Growth Des., 2023
|
|
8I77
 
 | | Crystal structure of horse spleen L-ferritin A115V mutant at -180deg Celsius. | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Maity, B, Tian, J, Abe, S, Ueno, T. | | Deposit date: | 2023-01-31 | | Release date: | 2023-03-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Atomic-level insights into a unique semi-clathrate hydrate formed in a confined environment of porous protein crystal.
Cryst.Growth Des., 2023
|
|
2ZEJ
 
 | |
1IYV
 
 | |
8I8Q
 
 | | Crystal structure of horse spleen L-ferritin H114A mutant at -180deg Celsius. | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Maity, B, Tian, J, Abe, S, Ueno, T. | | Deposit date: | 2023-02-05 | | Release date: | 2023-03-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Atomic-level insights into a unique semi-clathrate hydrate formed in a confined environment of porous protein crystal.
Cryst.Growth Des., 2023
|
|
4CT8
 
 | |
