5K1R
 
 | |
4Y19
 
 | | immune complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FS18_alpha, FS18_beta, ... | | Authors: | Beringer, D.X, Petersen, J, Reid, H.H, Rossjohn, J. | | Deposit date: | 2015-02-07 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | T cell receptor reversed polarity recognition of a self-antigen major histocompatibility complex.
Nat.Immunol., 16, 2015
|
|
5M96
 
 | |
5MLL
 
 | |
8TUB
 
 | | HLA B7:02 with HPNGYKSLSTL | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Littler, D.R, Rossjohn, J, Chaurasia, P, Petersen, J. | | Deposit date: | 2023-08-16 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CD8 + T-cell responses towards conserved influenza B virus epitopes across anatomical sites and age.
Nat Commun, 15, 2024
|
|
8QU1
 
 | | mt-LSU assembly intermediate in GTPBP8 knock-out cells, state 1 | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Valentin Gese, G, Cipullo, M, Rorbach, J, Hallberg, B.M. | | Deposit date: | 2023-10-13 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | GTPBP8 plays a role in mitoribosome formation in human mitochondria.
Nat Commun, 15, 2024
|
|
8QRL
 
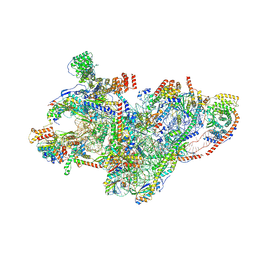 | | mt-SSU assembly intermediate in GTPBP8 knock-out cells, state 2 | | Descriptor: | 12S mitochondrial rRNA, 12S rRNA N4-methylcytidine (m4C) methyltransferase, 28S ribosomal protein S10, ... | | Authors: | Valentin Gese, G, Cipullo, M, Rorbach, J, Hallberg, B.M. | | Deposit date: | 2023-10-09 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | GTPBP8 plays a role in mitoribosome formation in human mitochondria.
Nat Commun, 15, 2024
|
|
8QRK
 
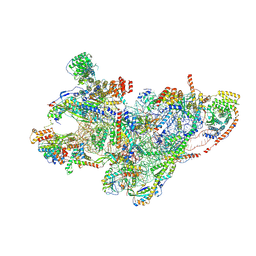 | | mt-SSU assembly intermediate in GTPBP8 knock-out cells, state 1 | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Valentin Gese, G, Cipullo, M, Rorbach, J, Hallberg, B.M. | | Deposit date: | 2023-10-09 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (6.69 Å) | | Cite: | GTPBP8 plays a role in mitoribosome formation in human mitochondria.
Nat Commun, 15, 2024
|
|
8QU5
 
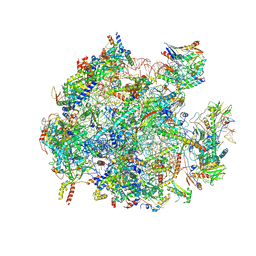 | | mt-LSU assembly intermediate in GTPBP8 knock-out cells, state 2 | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Valentin Gese, G, Cipullo, M, Rorbach, J, Hallberg, B.M. | | Deposit date: | 2023-10-13 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | GTPBP8 plays a role in mitoribosome formation in human mitochondria.
Nat Commun, 15, 2024
|
|
6UX2
 
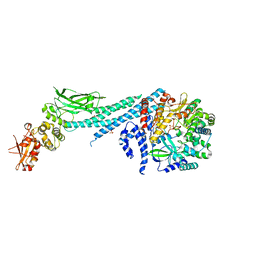 | | Crystal structure of ZIKV RdRp in complex with STAT2 | | Descriptor: | Nonstructural Protein 5, SULFATE ION, Signal transducer and activator of transcription 2, ... | | Authors: | Wang, B, Song, J. | | Deposit date: | 2019-11-06 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural basis for STAT2 suppression by flavivirus NS5.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8B1X
 
 | | Solution NMR structure of the single alpha helix peptide (P3-7)2 | | Descriptor: | P3-7_2 | | Authors: | Escobedo, A, Coles, M, Diercks, T, Garcia, J, Millet, O, Salvatella, X. | | Deposit date: | 2022-09-12 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A glutamine-based single alpha-helix scaffold to target globular proteins.
Nat Commun, 13, 2022
|
|
6UPY
 
 | | RNA polymerase II elongation complex with 5-guanidinohydantoin lesion in state 2E | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2019-10-18 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | RNA polymerase II stalls on oxidative DNA damage via a torsion-latch mechanism involving lone pair-pi and CH-pi interactions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UQ3
 
 | | RNA polymerase II elongation complex with 5-guanidinohydantoin lesion in state 5 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2019-10-18 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | RNA polymerase II stalls on oxidative DNA damage via a torsion-latch mechanism involving lone pair-pi and CH-pi interactions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7MSD
 
 | | Structure of EED bound to EEDi-6068 | | Descriptor: | (9aP,12aR)-4-(2,2-difluoropropyl)-12-{[(5-fluoro-2,3-dihydro-1-benzofuran-4-yl)methyl]amino}-7-(trifluoromethyl)-4,5-dihydro-3H-2,4,8,11,12a-pentaazabenzo[4,5]cycloocta[1,2,3-cd]inden-3-one, FORMIC ACID, Polycomb protein EED | | Authors: | Petrunak, E, Stuckey, J. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of EEDi-5273 as an Exceptionally Potent and Orally Efficacious EED Inhibitor Capable of Achieving Complete and Persistent Tumor Regression.
J.Med.Chem., 64, 2021
|
|
7MSB
 
 | | Structure of EED bound to EEDi-4259 | | Descriptor: | (9aM,12aS)-12-{[(5-fluoro-1-benzofuran-4-yl)methyl]amino}-7-(trifluoromethyl)-4,5-dihydro-3H-2,4,11,12a-tetraazabenzo[4,5]cycloocta[1,2,3-cd]inden-3-one, Polycomb protein EED | | Authors: | Petrunak, E, Stuckey, J. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of EEDi-5273 as an Exceptionally Potent and Orally Efficacious EED Inhibitor Capable of Achieving Complete and Persistent Tumor Regression.
J.Med.Chem., 64, 2021
|
|
6ROY
 
 | |
2XR6
 
 | | Crystal structure of the complex of the carbohydrate recognition domain of human DC-SIGN with pseudo trimannoside mimic. | | Descriptor: | 2-AZIDOETHANOL, CALCIUM ION, CD209 ANTIGEN, ... | | Authors: | Thepaut, M, Suitkeviciute, I, Sattin, S, Reina, J, Bernardi, A, Fieschi, F. | | Deposit date: | 2010-09-10 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Unique Dc-Sign Clustering Activity of a Small Glycomimetic: A Lesson for Ligand Design.
Acs Chem.Biol., 9, 2014
|
|
4RGH
 
 | |
8AN6
 
 | |
7R6A
 
 | |
8ASR
 
 | |
6UQ0
 
 | | RNA polymerase II elongation complex with 5-guanidinohydantoin lesion in state 4 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2019-10-18 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | RNA polymerase II stalls on oxidative DNA damage via a torsion-latch mechanism involving lone pair-pi and CH-pi interactions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UPX
 
 | | RNA polymerase II elongation complex with 5-guanidinohydantoin lesion in state 1 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2019-10-18 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | RNA polymerase II stalls on oxidative DNA damage via a torsion-latch mechanism involving lone pair-pi and CH-pi interactions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5K8Y
 
 | | Structure of the Mus musclus Langerin carbohydrate recognition domain | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION, GLYCEROL, ... | | Authors: | Loll, B, Aretz, J, Rademacher, C, Wahl, M.C. | | Deposit date: | 2016-05-31 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bacterial Polysaccharide Specificity of the Pattern Recognition Receptor Langerin Is Highly Species-dependent.
J. Biol. Chem., 292, 2017
|
|
8AST
 
 | |
