5J2Y
 
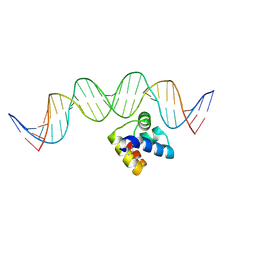 | | Molecular insight into the regulatory mechanism of the quorum-sensing repressor RsaL in Pseudomonas aeruginosa | | Descriptor: | DNA (26-MER), Regulatory protein | | Authors: | Zhao, J, Gan, J, Zhang, J, Kang, H, Kong, W, Zhu, M, Li, F, Song, Y, Qin, J, Liang, H. | | Deposit date: | 2016-03-30 | | Release date: | 2017-04-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Pseudomonas aeruginosa RsaL bound to promoter DNA reaffirms its role as a global regulator involved in quorum-sensing.
Nucleic Acids Res., 45, 2017
|
|
4WZJ
 
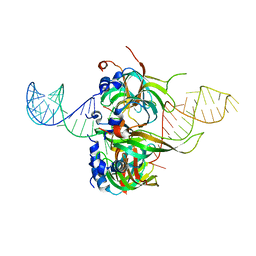 | | Spliceosomal U4 snRNP core domain | | Descriptor: | Small nuclear ribonucleoprotein E, Small nuclear ribonucleoprotein F, Small nuclear ribonucleoprotein G, ... | | Authors: | Leung, A.K.W, Nagai, K, Li, J. | | Deposit date: | 2014-11-19 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of the spliceosomal U4 snRNP core domain and its implication for snRNP biogenesis.
Nature, 473, 2011
|
|
4TNK
 
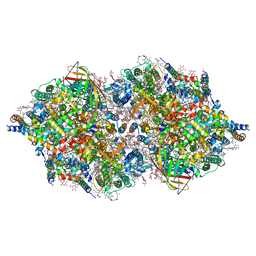 | | RT XFEL structure of Photosystem II 250 microsec after the third illumination at 5.2 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (5.2 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
7MRR
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor Leupeptin | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, LEUPEPTIN | | Authors: | Andi, B, Kumaran, D, Soares, A.S, Kreitler, D.F, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2021-05-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
8UPT
 
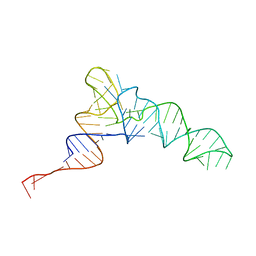 | | Candidatus Methanomethylophilus alvus tRNAPyl in A-site of ribosome | | Descriptor: | RNA (71-MER) | | Authors: | Krahn, N, Zhang, J, Melnikov, S.V, Tharp, J.M, Villa, A, Patel, A, Howard, R.J, Gabir, H, Patel, T.R, Stetefeld, J, Puglisi, J, Soll, D. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | tRNA shape is an identity element for an archaeal pyrrolysyl-tRNA synthetase from the human gut.
Nucleic Acids Res., 52, 2024
|
|
6PDZ
 
 | |
6OVO
 
 | | Crystal structure of the unliganded PG10 TCR | | Descriptor: | 1,2-ETHANEDIOL, Alpha Chain T-Cell Receptor PG10, Beta Chain T-Cell Receptor PG10, ... | | Authors: | Shahine, A, Rossjohn, J. | | Deposit date: | 2019-05-08 | | Release date: | 2019-11-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | A TCR beta-Chain Motif Biases toward Recognition of Human CD1 Proteins.
J Immunol., 203, 2019
|
|
8H10
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-10-19 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H13
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x2 Disulfide (G400C and V969C), Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-10-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H14
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x3 Disulfide (D414C and V969C), Locked-1 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H16
 
 | | Structure of SARS-CoV-1 Spike Protein (S/native) at pH 5.5, Open Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.35534 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H11
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H12
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x2 Disulfide (G400C and V969C), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.44681 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H0Y
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-112 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, LINOLEIC ACID, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H15
 
 | | Structure of SARS-CoV-1 Spike Protein (S/native) at pH 5.5, Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.14182 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H0X
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-1 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, LINOLEIC ACID, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
1DWS
 
 | | PHOTOLYZED CARBONMONOXY MYOGLOBIN (HORSE HEART) | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Chu, K, Vojtechovsky, J, McMahon, B.H, Sweet, R.M, Berendzen, J, Schlichting, I. | | Deposit date: | 1999-12-11 | | Release date: | 2000-03-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of a New Ligand Binding Intermediate in Wildtype Carbonmonoxy Myoglobin
Nature, 403, 2000
|
|
1DWR
 
 | | MYOGLOBIN (HORSE HEART) WILD-TYPE COMPLEXED WITH CO | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Chu, K, Vojtechovsky, J, McMahon, B.H, Sweet, R.M, Berendzen, J, Schlichting, I. | | Deposit date: | 1999-12-11 | | Release date: | 2000-03-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of a New Ligand Binding Intermediate in Wildtype Carbonmonoxy Myoglobin
Nature, 403, 2000
|
|
7A5V
 
 | | CryoEM structure of a human gamma-aminobutyric acid receptor, the GABA(A)R-beta3 homopentamer, in complex with histamine and megabody Mb25 in lipid nanodisc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Nakane, T, Kotecha, A, Sente, A, Yamashita, K, McMullan, G, Masiulis, S, Brown, P.M.G.E, Grigoras, I.T, Malinauskaite, L, Malinauskas, T, Miehling, J, Yu, L, Karia, D, Pechnikova, E.V, de Jong, E, Keizer, J, Bischoff, M, McCormack, J, Tiemeijer, P, Hardwick, S.W, Chirgadze, D.Y, Murshudov, G, Aricescu, A.R, Scheres, S.H.W. | | Deposit date: | 2020-08-22 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (1.7 Å) | | Cite: | Single-particle cryo-EM at atomic resolution.
Nature, 587, 2020
|
|
6TFY
 
 | | Crystal Structure of EGFR T790M/V948R in Complex with Covalent Pyrrolopyrimidine 18c | | Descriptor: | Epidermal growth factor receptor, SULFATE ION, ~{N}-[5-[4-[[3-chloranyl-4-(pyridin-2-ylmethoxy)phenyl]amino]-7~{H}-pyrrolo[2,3-d]pyrimidin-5-yl]-2-(3-oxidanylpropoxy)phenyl]propanamide | | Authors: | Niggenaber, J, Mueller, M.P, Rauh, D. | | Deposit date: | 2019-11-14 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Targeting Her2-insYVMA with Covalent Inhibitors-A Focused Compound Screening and Structure-Based Design Approach.
J.Med.Chem., 63, 2020
|
|
8Q4S
 
 | |
7MNG
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor VBY-825 (Partial Occupancy) | | Descriptor: | (2R,3S)-N-cyclopropyl-3-{[(2R)-3-(cyclopropylmethanesulfonyl)-2-{[(1S)-2,2,2-trifluoro-1-(4-fluorophenyl)ethyl]amino}propanoyl]amino}-2-hydroxypentanamide (non-preferred name), 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Andi, B, Kumaran, D, Soares, A.S, Kreitler, D.F, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2021-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
4X48
 
 | | Crystal structure of GluR2 ligand-binding core | | Descriptor: | GLUTAMIC ACID, Glutamate receptor 2, N-{(3S,4S)-4-[4-(5-cyanothiophen-2-yl)phenoxy]tetrahydrofuran-3-yl}propane-2-sulfonamide, ... | | Authors: | Pandit, J. | | Deposit date: | 2014-12-02 | | Release date: | 2015-05-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The Discovery and Characterization of the alpha-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA) Receptor Potentiator N-{(3S,4S)-4-[4-(5-Cyano-2-thienyl)phenoxy]tetrahydrofuran-3-yl}propane-2-sulfonamide (PF-04958242).
J.Med.Chem., 58, 2015
|
|
1DWT
 
 | | Photorelaxed horse heart MYOGLOBIN CO complex | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Chu, K, Vojtechovsky, J, McMahon, B.H, Sweet, R.M, Berendzen, J, Schlichting, I. | | Deposit date: | 1999-12-12 | | Release date: | 2000-03-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of a New Ligand Binding Intermediate in Wildtype Carbonmonoxy Myoglobin
Nature, 403, 2000
|
|
7A4M
 
 | | Cryo-EM structure of mouse heavy-chain apoferritin at 1.22 A | | Descriptor: | FE (III) ION, Ferritin heavy chain, ZINC ION | | Authors: | Nakane, T, Kotecha, A, Sente, A, Yamashita, K, McMullan, G, Masiulis, S, Brown, P.M.G.E, Grigoras, I.T, Malinauskaite, L, Malinauskas, T, Miehling, J, Yu, L, Karia, D, Pechnikova, E.V, de Jong, E, Keizer, J, Bischoff, M, McCormack, J, Tiemeijer, P, Hardwick, S.W, Chirgadze, D.Y, Murshudov, G, Aricescu, A.R, Scheres, S.H.W. | | Deposit date: | 2020-08-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (1.22 Å) | | Cite: | Single-particle cryo-EM at atomic resolution.
Nature, 587, 2020
|
|
