3J8A
 
 | | Structure of the F-actin-tropomyosin complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | von der Ecken, J, Mueller, M, Lehman, W, Manstein, J.M, Penczek, A.P, Raunser, S. | | Deposit date: | 2014-10-08 | | Release date: | 2014-12-10 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the F-actin--tropomyosin complex.
Nature, 519, 2015
|
|
4AUK
 
 | | Crystal structure of C2498 2'-O-ribose methyltransferase RlmM from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Punekar, A.S, Shepherd, T.R, Liljeruhm, J, Forster, A.C, Selmer, M. | | Deposit date: | 2012-05-18 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Rlmm, the 2'O-Ribose Methyltransferase for C2498 of Escherichia Coli 23S Rrna.
Nucleic Acids Res., 40, 2012
|
|
4B7N
 
 | | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE, ... | | Authors: | van der Vries, E, Vachieri, S.G, Xiong, X, Liu, J, Collins, P.J, Walker, P.A, Haire, L.F, Hay, A.J, Schutten, M, Osterhaus, A.D.M.E, Martin, S.R, Boucher, C.A.B, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2012-08-21 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis
Plos Pathog., 8, 2012
|
|
3J7N
 
 | | Virus model of brome mosaic virus (second half data set) | | Descriptor: | Capsid protein | | Authors: | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
6DE3
 
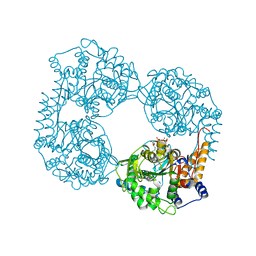 | | Crystal structure of the double mutant (R39Q/D52N) of the full-length NT5C2 in the active state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cytosolic purine 5'-nucleotidase, MAGNESIUM ION, ... | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L. | | Deposit date: | 2018-05-11 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
5W3X
 
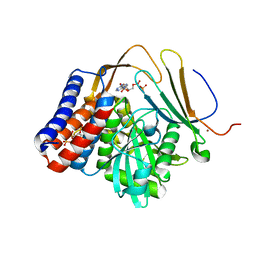 | | Crystal structure of PopP2 in complex with IP6, AcCoA and the WRKY domain of RRS1-R . | | Descriptor: | ACETYL COENZYME *A, Disease resistance protein RRS1, GLYCEROL, ... | | Authors: | Zhang, Z.M, Gao, L, Song, J. | | Deposit date: | 2017-06-08 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of host substrate acetylation by a YopJ family effector.
Nat Plants, 3, 2017
|
|
4B73
 
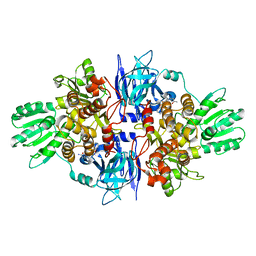 | | Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function | | Descriptor: | (2S)-4-amino-N-[(1R)-1-(4-chloro-2-fluoro-3-phenoxyphenyl)propyl]-4-oxobutan-2-aminium, NON-STRUCTURAL PROTEIN 4A, SERINE PROTEASE NS3 | | Authors: | Saalau-Bethell, S.M, Woodhead, A.J, Chessari, G, Carr, M.G, Coyle, J, Graham, B, Hiscock, S.D, Murray, C.W, Pathuri, P, Rich, S.J, Richardson, C.J, Williams, P.A, Jhoti, H. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of an Allosteric Mechanism for the Regulation of Hcv Ns3 Protein Function.
Nat.Chem.Biol., 8, 2012
|
|
1E93
 
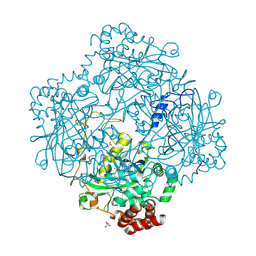 | | High resolution structure and biochemical properties of a recombinant catalase depleted in iron | | Descriptor: | ACETATE ION, CATALASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Andreoletti, P, Sainz, G, Jaquinod, M, Gagnon, J, Jouve, H.M. | | Deposit date: | 2000-10-05 | | Release date: | 2000-10-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-Resolution Structure and Biochemical Properties of a Recombinant Proteus Mirabilis Catalase Depleted in Iron.
Proteins: Struct.,Funct., Genet., 50, 2003
|
|
3HA2
 
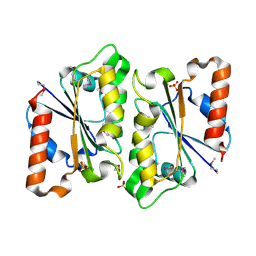 | | Crystal Structure of Protein (NADPH-quinone reductase) from P.pentosaceus, Northeast Structural Genomics Consortium Target PtR24A | | Descriptor: | DI(HYDROXYETHYL)ETHER, NADPH-quinone reductase, SULFATE ION | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Maglaqui, M, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-04-30 | | Release date: | 2009-05-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Northeast Structural Genomics Consortium Target PtR24A
To be Published
|
|
5W4A
 
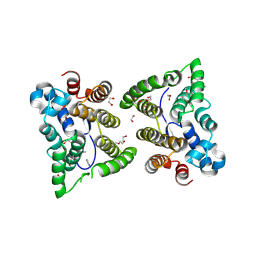 | | C. japonica N-domain | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IMIDAZOLE, ... | | Authors: | Aoki, S.T, Bingman, C.A, Kimble, J. | | Deposit date: | 2017-06-09 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | C. elegans germ granules require both assembly and localized regulators for mRNA repression.
Nat Commun, 12, 2021
|
|
3HB7
 
 | | The Crystal Structure of an Isochorismatase-like Hydrolase from Alkaliphilus metalliredigens to 2.3A | | Descriptor: | AMMONIUM ION, Isochorismatase hydrolase, SODIUM ION | | Authors: | Stein, A.J, Xu, X, Cui, H, Ng, J, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-04 | | Release date: | 2009-07-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of an Isochorismatase-like Hydrolase from Alkaliphilus metalliredigens to 2.3A
To be Published
|
|
5W4Y
 
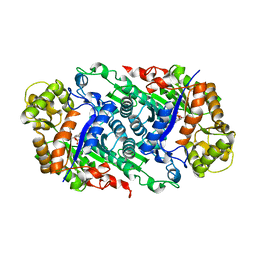 | | Crystal Structure of Riboflavin Lyase (RcaE) with cofactor FMN | | Descriptor: | FLAVIN MONONUCLEOTIDE, Riboflavin Lyase | | Authors: | Bhandari, D.M, Chakrabarty, Y, Zhao, B, Wood, J, Li, P, Begley, T.P. | | Deposit date: | 2017-06-13 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cannibalism Among the Flavins: a Novel C-N Bond Cleavage in Riboflavin Catabolism Mediated by Flavin-Generated Superoxide Radical
To be Published
|
|
4AV6
 
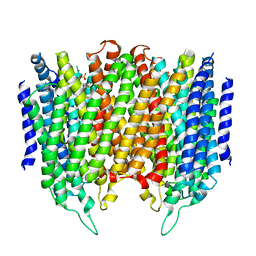 | | Crystal structure of Thermotoga maritima sodium pumping membrane integral pyrophosphatase at 4 A in complex with phosphate and magnesium | | Descriptor: | K(+)-STIMULATED PYROPHOSPHATE-ENERGIZED SODIUM PUMP, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kajander, T, Kellosalo, J, Kogan, K, Pokharel, K, Goldman, A. | | Deposit date: | 2012-05-23 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The Structure and Catalytic Cycle of a Sodium-Pumping Pyrophosphatase.
Science, 337, 2012
|
|
6HGQ
 
 | | Crystal Structure of Human APRT wild type in complex with Hypoxanthine, PRPP and Mg2+ | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, Adenine phosphoribosyltransferase, GLYCEROL, ... | | Authors: | Nioche, P, Huyet, J, Ozeir, M. | | Deposit date: | 2018-08-23 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for substrate selectivity and nucleophilic substitution mechanisms in human adenine phosphoribosyltransferase catalyzed reaction.
J.Biol.Chem., 294, 2019
|
|
4B2G
 
 | | Crystal Structure of an Indole-3-Acetic Acid Amido Synthase from Vitis vinifera Involved in Auxin Homeostasis | | Descriptor: | GH3-1 AUXIN CONJUGATING ENZYME, MALONATE ION, [(2S,3R,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl] 2-(1H-indol-3-yl)ethyl hydrogen phosphate | | Authors: | Peat, T.S, Bottcher, C, Newman, J, Lucent, D, Cowieson, N, Davies, C. | | Deposit date: | 2012-07-16 | | Release date: | 2012-12-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of an Indole-3-Acetic Acid Amido Synthetase from Grapevine Involved in Auxin Homeostasis.
Plant Cell, 24, 2012
|
|
4B4P
 
 | | Crystal Structure of the lectin domain of F18 fimbrial adhesin FedF. | | Descriptor: | BROMIDE ION, F18 FIMBRIAL ADHESIN AC, SULFATE ION | | Authors: | Moonens, K, Bouckaert, J, Coddens, A, Tran, T, Panjikar, S, De Kerpel, M, Cox, E, Remaut, H, De Greve, H. | | Deposit date: | 2012-07-31 | | Release date: | 2012-08-15 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insight in Histo-Blood Group Binding by the F18 Fimbrial Adhesin Fedf.
Mol.Microbiol., 86, 2012
|
|
3EZH
 
 | |
1EBE
 
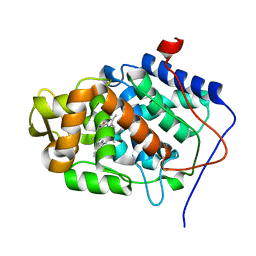 | | Laue diffraction study on the structure of cytochrome c peroxidase compound I | | Descriptor: | CYTOCHROME C PEROXIDASE, OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fulop, V, Phizackerley, R.P, Soltis, S.M, Clifton, I.J, Wakatsuki, S, Erman, J.E, Hajdu, J, Edwards, S.L. | | Deposit date: | 2001-07-25 | | Release date: | 2001-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Laue Diffraction Study on the Structure of Cytochrome C Peroxidase Compound I
Structure, 2, 1994
|
|
1KNB
 
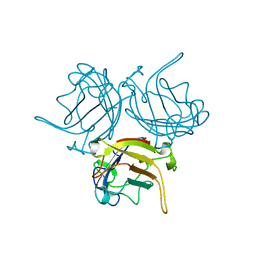 | |
2N2E
 
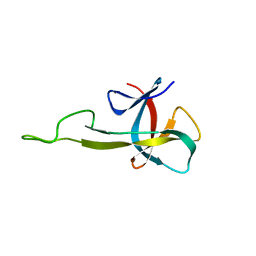 | | NMR solution structure of the C-terminal domain of NisI, a lipoprotein from Lactococcus lactis which confers immunity against nisin | | Descriptor: | Nisin immunity protein | | Authors: | Hacker, C, Christ, N.A, Korn, S, Duchardt-Ferner, E, Hellmich, U.A, Duesterhus, S, Koetter, P, Entian, K, Woehnert, J. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of the Lantibiotic Immunity Protein NisI and Its Interactions with Nisin.
J.Biol.Chem., 290, 2015
|
|
1ELM
 
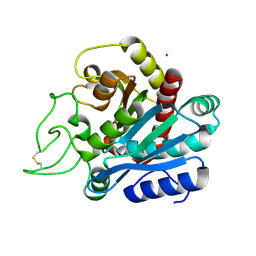 | | CADMIUM-SUBSTITUTED BOVINE PACREATIC CARBOXYPEPTIDASE A (ALFA-FORM) AT PH 5.5 AND 2 MM CHLORIDE IN MONOCLINIC CRYSTAL FORM. | | Descriptor: | CADMIUM ION, CARBOXYPEPTIDASE A | | Authors: | Jensen, F, Bukrinsky, T, Bjerrum, J, Larsen, S. | | Deposit date: | 2000-03-14 | | Release date: | 2002-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three high-resolution crystal structures of cadmium-substituted carboxypeptidase A provide insight into the enzymatic function
J.BIOL.INORG.CHEM., 7, 2002
|
|
4B7Q
 
 | | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE, ... | | Authors: | Liu, J, van der Vries, E, Vachieri, S.G, Xiong, X, Collins, P.J, Walker, P.A, Haire, L.F, Hay, A.J, Schutten, M, Osterhaus, A.D.M.E, Martin, S.R, Boucher, C.A.B, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2012-08-21 | | Release date: | 2012-10-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.728 Å) | | Cite: | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis
Plos Pathog., 8, 2012
|
|
6HHT
 
 | | Echovirus 18 Open particle without two pentamers | | Descriptor: | Echovirus 18 capsid protein 1, Echovirus 18 capsid protein 2, Echovirus 18 capsid protein 3 | | Authors: | Buchta, D, Fuzik, T, Hrebik, D, Levdansky, Y, Moravcova, J, Plevka, P. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-20 | | Last modified: | 2019-04-10 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Enterovirus particles expel capsid pentamers to enable genome release.
Nat Commun, 10, 2019
|
|
1EE3
 
 | | Cadmium-substituted bovine pancreatic carboxypeptidase A (alfa-form) at pH 7.5 and 2 mM chloride in monoclinic crystal form | | Descriptor: | CADMIUM ION, PROTEIN (CARBOXYPEPTIDASE A) | | Authors: | Jensen, F, Bukrinsky, T, Bjerrum, J, Larsen, S. | | Deposit date: | 2000-01-30 | | Release date: | 2002-06-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Three high-resolution crystal structures of cadmium-substituted carboxypeptidase A provide insight into the enzymatic function
J.BIOL.INORG.CHEM., 7, 2002
|
|
3F23
 
 | | Crystal structure of Zalpha in complex with d(CGGCCG) | | Descriptor: | DNA (5'-D(*DTP*DCP*DGP*DGP*DCP*DCP*DG)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Ha, S.C, Choi, J, Kim, K.K. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structures of non-CG-repeat Z-DNAs co-crystallized with the Z-DNA-binding domain, hZ{alpha}ADAR1
Nucleic Acids Res., 37, 2009
|
|
