5VVX
 
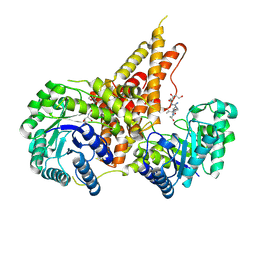 | | Structural Investigations of the Substrate Specificity of Human O-GlcNAcase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lamin B1, Protein O-GlcNAcase | | Authors: | Li, B, Jiang, J, Li, H, Hu, C.-W. | | Deposit date: | 2017-05-21 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the substrate binding adaptability and specificity of human O-GlcNAcase.
Nat Commun, 8, 2017
|
|
4CV9
 
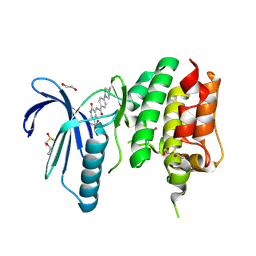 | | MPS1 kinase with 3-aminopyridin-2-one inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-Methylpiperazin-1-yl)-N-(2-oxo-5-(pyridin-4-yl)-1,2-dihydropyridin-3-yl)benzamide, DIMETHYL SULFOXIDE, ... | | Authors: | Fearon, D, Bavetsias, V, Bayliss, R, Schmitt, J, Westwood, I.M, vanMontfort, R.L.M, Jones, K. | | Deposit date: | 2014-03-24 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein Kinase Selectivity of a 3-Aminopyridin-2- One Based Fragment Library, Identification of 3-Amino-5-(Pyridin-4-Yl)Pyridin-2(1H)-One as a Novel Scaffold for Mps1 Inhibition
To be Published
|
|
5VHP
 
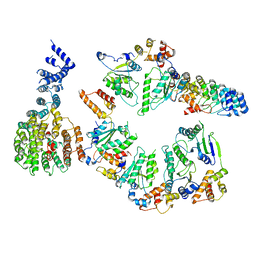 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 10, 26S proteasome non-ATPase regulatory subunit 2, 26S proteasome regulatory subunit 10B, ... | | Authors: | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
4D47
 
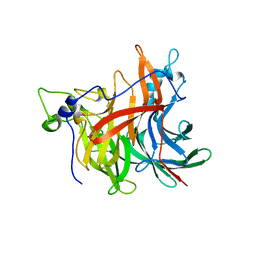 | | X-ray structure of the levansucrase from Erwinia amylovora | | Descriptor: | LEVANSUCRASE, alpha-D-glucopyranose, beta-D-fructofuranose | | Authors: | Wuerges, J, Caputi, L, Cianci, M, Benini, S. | | Deposit date: | 2014-10-27 | | Release date: | 2015-08-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | The Crystal Structure of Erwinia Amylovora Levansucrase Provides a Snapshot of the Products of Sucrose Hydrolysis Trapped Into the Active Site.
J.Struct.Biol., 191, 2015
|
|
5VHZ
 
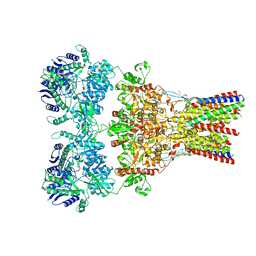 | | GluA2-2xGSG1L bound to L-Quisqualate | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 2,Germ cell-specific gene 1-like protein | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2017-04-13 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Structural Bases of Desensitization in AMPA Receptor-Auxiliary Subunit Complexes.
Neuron, 94, 2017
|
|
3HYM
 
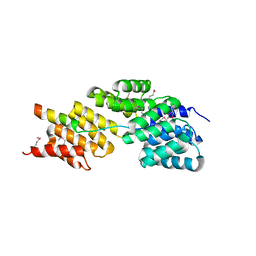 | | Insights into Anaphase Promoting Complex TPR subdomain assembly from a CDC26-APC6 structure | | Descriptor: | Anaphase-promoting complex subunit CDC26, Cell division cycle protein 16 homolog | | Authors: | Wang, J, Dye, B.T, Rajashankar, K.R, Kurinov, I, Schulman, B.A. | | Deposit date: | 2009-06-22 | | Release date: | 2009-08-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into anaphase promoting complex TPR subdomain assembly from a CDC26-APC6 structure.
Nat.Struct.Mol.Biol., 16, 2009
|
|
4CQP
 
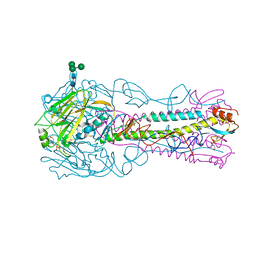 | | Crystal Structure of H5 (VN1194) Ser227Asn/Gln196Arg Mutant Haemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Xiong, X, Xiao, H, Martin, S.R, Coombs, P.J, Liu, J, Collins, P.J, Vachieri, S.G, Walker, P.A, Lin, Y.P, McCauley, J.W, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2014-02-21 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Enhanced Human Receptor Binding by H5 Haemagglutinins.
Virology, 456, 2014
|
|
6JT8
 
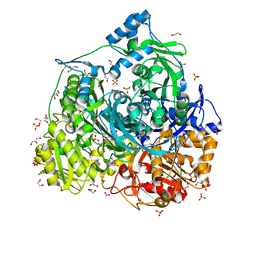 | | Crystal structure of 450-451_deletion mutant of FGAM Synthetase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Sharma, N, Ahalawat, N, Sandhu, P, Mondal, J, Anand, R. | | Deposit date: | 2019-04-10 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of allosteric switches and adaptor domains in long-distance cross-talk and transient tunnel formation.
Sci Adv, 6, 2020
|
|
3I3G
 
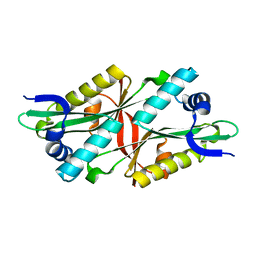 | | Crystal Structure of Trypanosoma brucei N-acetyltransferase (Tb11.01.2886) at 1.86A | | Descriptor: | N-acetyltransferase | | Authors: | Qiu, W, Wernimont, A.K, Marino, K, Zhang, A.Z, Ma, D, Lin, Y.H, Mackenzie, F, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, J Ferguson, M.A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-30 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure Trypanosoma brucei N-acetyltransferase (Tb11.01.2886) at 1.86A
To be Published
|
|
1KKA
 
 | |
5VL3
 
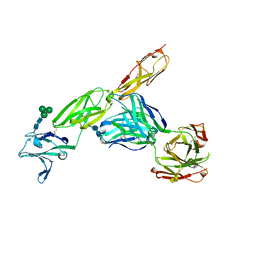 | | CD22 d1-d3 in complex with therapeutic Fab Epratuzumab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, B-cell receptor CD22, Epratuzumab Fab Heavy Chain, ... | | Authors: | Sicard, T, Ereno-Orbea, J, Julien, J.P. | | Deposit date: | 2017-04-24 | | Release date: | 2017-10-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular basis of human CD22 function and therapeutic targeting.
Nat Commun, 8, 2017
|
|
3CL2
 
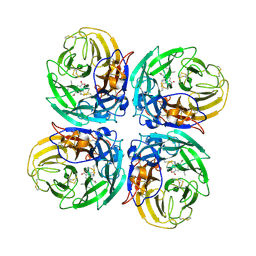 | | N1 Neuraminidase N294S + Oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, Neuraminidase | | Authors: | Collins, P, Haire, L.F, Lin, Y.P, Liu, J, Russell, R.J, Walker, P.A, Skehel, J.J, Martin, S.R, Hay, A.J, Gamblin, S.J. | | Deposit date: | 2008-03-18 | | Release date: | 2008-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.538 Å) | | Cite: | Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants.
Nature, 453, 2008
|
|
5VEX
 
 | | Structure of the human GLP-1 receptor complex with NNC0640 | | Descriptor: | 4-{[(4-cyclohexylphenyl){[3-(methylsulfonyl)phenyl]carbamoyl}amino]methyl}-N-(1H-tetrazol-5-yl)benzamide, Glucagon-like peptide 1 receptor, Endolysin chimera | | Authors: | Song, G, Yang, D, Wang, Y, Graaf, C.D, Zhou, Q, Jiang, S, Liu, K, Cai, X, Dai, A, Lin, G, Liu, D, Wu, F, Wu, Y, Zhao, S, Ye, L, Han, G.W, Lau, J, Wu, B, Hanson, M.A, Liu, Z.-J, Wang, M.-W, Stevens, R.C. | | Deposit date: | 2017-04-05 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators.
Nature, 546, 2017
|
|
6C9W
 
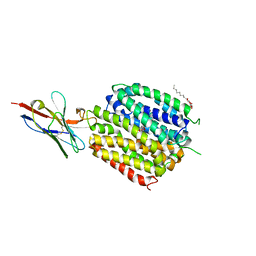 | | Crystal Structure of a ligand bound LacY/Nanobody Complex | | Descriptor: | 4-nitrophenyl alpha-D-galactopyranoside, Lactose permease, Nanobody9047, ... | | Authors: | Kumar, H, Finer-Moore, J.S, Jiang, X, Smirnova, I, Kasho, V, Pardon, E, Steyaert, J, Kaback, H.R, Stroud, R.M. | | Deposit date: | 2018-01-29 | | Release date: | 2018-08-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of a ligand-bound LacY-Nanobody Complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2O2H
 
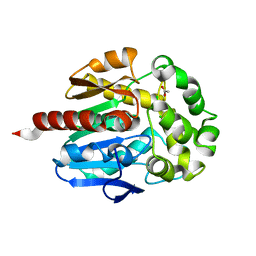 | | Crystal structure of haloalkane dehalogenase Rv2579 from Mycobacterium tuberculosis complexed with 1,2-dichloroethane | | Descriptor: | 1,2-DICHLOROETHANE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Mazumdar, P.A, Hulecki, J, Cherney, M.M, Garen, C.R, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-11-29 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of haloalkane dehalogenase Rv2579 from Mycobacterium tuberculosis complexed with 1,2-dichloroethane
TO BE PUBLISHED
|
|
5VHM
 
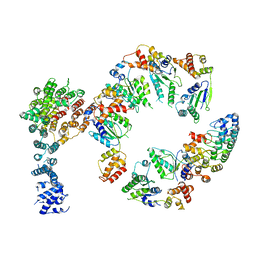 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 10, 26S proteasome non-ATPase regulatory subunit 2, 26S proteasome regulatory subunit 10B, ... | | Authors: | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
3D7G
 
 | |
5VNO
 
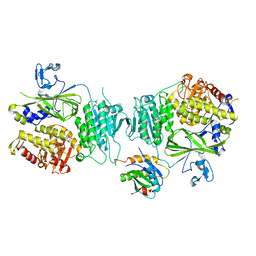 | | Crystal structure of Sec23a/Sec24a/Sec22 | | Descriptor: | Protein transport protein Sec23A, Protein transport protein Sec24A, Vesicle-trafficking protein SEC22b, ... | | Authors: | Ma, W, Goldberg, J. | | Deposit date: | 2017-05-01 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.905 Å) | | Cite: | ER retention is imposed by COPII protein sorting and attenuated by 4-phenylbutyrate.
Elife, 6, 2017
|
|
4D2Q
 
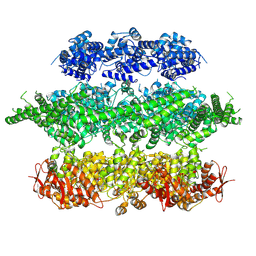 | | Negative-stain electron microscopy of E. coli ClpB mutant E432A (BAP form bound to ClpP) | | Descriptor: | CLPB | | Authors: | Carroni, M, Kummer, E, Oguchi, Y, Clare, D.K, Wendler, P, Sinning, I, Kopp, J, Mogk, A, Bukau, B, Saibil, H.R. | | Deposit date: | 2014-05-12 | | Release date: | 2014-06-04 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Head-to-Tail Interactions of the Coiled-Coil Domains Regulate Clpb Activity and Cooperation with Hsp70 in Protein Disaggregation.
Elife, 3, 2014
|
|
3HNP
 
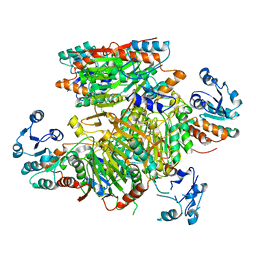 | | Crystal Structure of an Oxidoreductase from Bacillus cereus. Northeast Structural Genomics Consortium target id BcR251 | | Descriptor: | Oxidoreductase | | Authors: | Seetharaman, J, Su, M, Sahdev, S, Janjua, H, Xiao, R, Ciccosanti, C, Foote, E.L, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-05-31 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of an Oxidoreductase from Bacillus cereus. Northeast Structural Genomics Consortium target id BcR251
TO BE PUBLISHED
|
|
6C5W
 
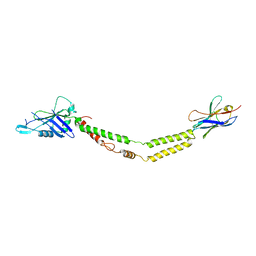 | | Crystal structure of the mitochondrial calcium uniporter | | Descriptor: | CALCIUM ION, calcium uniporter, nanobody | | Authors: | Fan, C, Fan, M, Fastman, N, Zhang, J, Feng, L. | | Deposit date: | 2018-01-17 | | Release date: | 2018-07-11 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (3.10010242 Å) | | Cite: | X-ray and cryo-EM structures of the mitochondrial calcium uniporter.
Nature, 559, 2018
|
|
1GB6
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-06-26 | | Release date: | 2000-07-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of surface hydrophobic residues in the conformational stability of human lysozyme at three different positions.
Biochemistry, 39, 2000
|
|
3HR1
 
 | | Discovery of novel inhibitors of PDE10A | | Descriptor: | 2-{[4-(1-methyl-4-pyridin-4-yl-1H-pyrazol-3-yl)phenoxy]methyl}quinoline, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Pandit, J, Marr, E.S. | | Deposit date: | 2009-06-08 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Discovery of a Novel Class of Phosphodiesterase 10A Inhibitors and Identification of Clinical Candidate 2-[4-(1-Methyl-4-pyridin-4-yl-1H-pyrazol-3-yl)-phenoxymethyl]-quinoline (PF-2545920) for the Treatment of Schizophrenia
J.Med.Chem., 52, 2009
|
|
4CP9
 
 | | Crystal structure OF lecA lectin complexed with a divalent galactoside at 1.65 angstrom | | Descriptor: | (4S)-N-ethyl-4-{[N-methyl-3-(1-{2-[(4-sulfanylbenzoyl)amino]ethyl}-1H-1,2,3-triazol-4-yl)-L-alanyl]amino}-L-prolinamide, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Topin, J, Varrot, A, Imberty, A, Wissinger, N. | | Deposit date: | 2014-02-04 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Leca Ligand Identified from a Galactoside-Conjugate Array Inhibits Host Cell Invasion by Pseudomonas Aeruginosa.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3DNN
 
 | | Molecular structure for the HIV-1 gp120 trimer in the unliganded state | | Descriptor: | HIV-1 envelope glycoprotein gp120 | | Authors: | Borgnia, M.J, Liu, J, Bartesaghi, A, Sapiro, G, Subramaniam, S. | | Deposit date: | 2008-07-02 | | Release date: | 2008-08-19 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Molecular architecture of native HIV-1 gp120 trimers.
Nature, 455, 2008
|
|
