6DEU
 
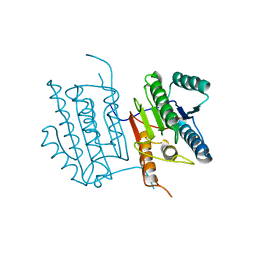 | | Human caspase-6 A109T | | Descriptor: | Caspase-6 | | Authors: | Tubeleviciute-Aydin, A, Beautrait, A, Lynham, J, Sharma, G, Gorelik, A, Deny, L.J, Soya, N, Lukacs, G.L, Nagar, B, Marinier, A, LeBlanc, A.C. | | Deposit date: | 2018-05-13 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Identification of Allosteric Inhibitors against Active Caspase-6.
Sci Rep, 9, 2019
|
|
4B5H
 
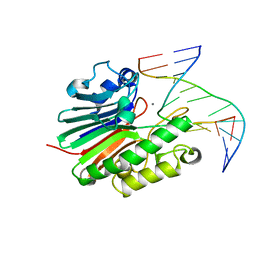 | | Substate bound inactive mutant of Neisseria AP endonuclease in presence of metal ions | | Descriptor: | 5'-D(*CP*GP*AP*TP*GP*GP*GP*TP*AP*GP*CP)-3', 5'-D(*GP*CP*TP*AP*CP*3DRP*CP*AP*TP*CP*GP)-3', MANGANESE (II) ION, ... | | Authors: | Lu, D, Silhan, J, MacDonald, J.T, Carpenter, E.P, Jensen, K, Tang, C.M, Baldwin, G.S, Freemont, P.S. | | Deposit date: | 2012-08-03 | | Release date: | 2012-10-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural basis for the recognition and cleavage of abasic DNA in Neisseria meningitidis.
Proc. Natl. Acad. Sci. U.S.A., 109, 2012
|
|
4B5M
 
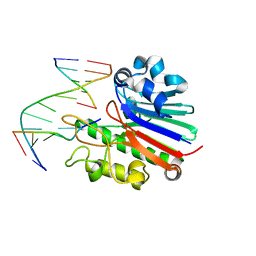 | | Neisseria AP endonuclease bound to the substrate with a cytosine orphan base | | Descriptor: | 5'-D(*3DRP*CP*AP*TP*CP*GP)-3', 5'-D(*CP*GP*AP*TP*GP*CP*GP*TP*AP*GP*CP)-3', 5'-D(*GP*CP*TP*AP*CP)-3', ... | | Authors: | Lu, D, Silhan, J, MacDonald, J.T, Carpenter, E.P, Jensen, K, Tang, C.M, Baldwin, G.S, Freemont, P.S. | | Deposit date: | 2012-08-06 | | Release date: | 2012-10-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.758 Å) | | Cite: | Structural basis for the recognition and cleavage of abasic DNA in Neisseria meningitidis.
Proc. Natl. Acad. Sci. U.S.A., 109, 2012
|
|
3HP4
 
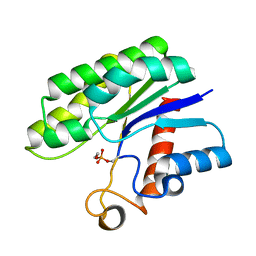 | | Crystal structure of psychrotrophic esterase EstA from Pseudoalteromonas sp. 643A inhibited by monoethylphosphonate | | Descriptor: | GDSL-esterase | | Authors: | Brzuszkiewicz, A, Nowak, E, Dauter, Z, Dauter, M, Cieslinski, H, Kur, J. | | Deposit date: | 2009-06-03 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of EstA esterase from psychrotrophic Pseudoalteromonas sp. 643A covalently inhibited by monoethylphosphonate.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3HP7
 
 | | Putative hemolysin from Streptococcus thermophilus. | | Descriptor: | ETHANOL, Hemolysin, putative | | Authors: | Osipiuk, J, Hatzos, C, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-06-03 | | Release date: | 2009-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | X-ray crystal structure of putative hemolysin from Streptococcus thermophilus.
To be Published
|
|
4B22
 
 | | Unprecedented sculpting of DNA at abasic sites by DNA glycosylase homolog Mag2 | | Descriptor: | 5'-D(*CP*GP*AP*TP*GP*GP*GP*TP*AP*GP*CP)-3', 5'-D(*GP*CP*TP*AP*CP*(3DR)P*CP*AP*TP*CP*GP)-3', MAG2, ... | | Authors: | Dalhus, B, Nilsen, L, Korvald, H, Huffman, J, Forstrom, R.J, McMurray, C.T, Alseth, I, Tainer, J.A, Bjoras, M. | | Deposit date: | 2012-07-12 | | Release date: | 2013-01-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sculpting of DNA at Abasic Sites by DNA Glycosylase Homolog Mag2.
Structure, 21, 2013
|
|
4BIJ
 
 | | Threading model of T7 large terminase | | Descriptor: | DNA MATURASE B | | Authors: | Dauden, M.I, Martin-Benito, J, Sanchez-Ferrero, J.C, Pulido-Cid, M, Valpuesta, J.M, Carrascosa, J.L. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Large Terminase Conformational Change Induced by Connector Binding in Bacteriophage T7
J.Biol.Chem., 288, 2013
|
|
5VRF
 
 | | CryoEM Structure of the Zinc Transporter YiiP from helical crystals | | Descriptor: | Cadmium and zinc efflux pump FieF, ZINC ION | | Authors: | Coudray, N, Lopez-Redondo, M, Zhang, Z, Alexopoulos, J, Stokes, D.L, Transcontinental EM Initiative for Membrane Protein Structure (TEMIMPS) | | Deposit date: | 2017-05-10 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for the alternating access mechanism of the cation diffusion facilitator YiiP.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6I19
 
 | | Crystal structure of Chlamydomonas reinhardtii thioredoxin h1 | | Descriptor: | Thioredoxin H-type | | Authors: | Lemaire, S.D, Tedesco, D, Crozet, P, Michelet, L, Fermani, S, Zaffagnini, M, Henri, J. | | Deposit date: | 2018-10-27 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.378 Å) | | Cite: | Crystal Structure of Chloroplastic Thioredoxin f2 fromChlamydomonas reinhardtiiReveals Distinct Surface Properties.
Antioxidants (Basel), 7, 2018
|
|
6DRJ
 
 | | Structure of TRPM2 ion channel receptor by single particle electron cryo-microscopy, ADPR/Ca2+ bound state | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CALCIUM ION, Transient receptor potential cation channel, ... | | Authors: | Du, J, Lu, W, Huang, Y, Winkler, P, Sun, W. | | Deposit date: | 2018-06-12 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Architecture of the TRPM2 channel and its activation mechanism by ADP-ribose and calcium.
Nature, 562, 2018
|
|
3CTF
 
 | | Crystal structure of oxidized GRX2 | | Descriptor: | Glutaredoxin-2 | | Authors: | Yu, J, Teng, Y.B, Zhou, C.Z. | | Deposit date: | 2008-04-14 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the different activities of yeast Grx1 and Grx2.
Biochim.Biophys.Acta, 1804, 2010
|
|
3HXJ
 
 | | Crystal Structure of Pyrrolo-quinoline quinone (PQQ_DH) from Methanococcus maripaludis, Northeast Structural Genomics Consortium Target MrR86 | | Descriptor: | Pyrrolo-quinoline quinone, SULFATE ION | | Authors: | Forouhar, F, Chen, Y, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Foote, E.L, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-20 | | Release date: | 2009-08-25 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Northeast Structural Genomics Consortium Target MrR86
To be Published
|
|
5VJI
 
 | | Crystal structure of the CLOCK Transcription Domain Exon19 in Complex with a Repressor | | Descriptor: | CLOCK-interacting pacemaker, Circadian locomoter output cycles protein kaput | | Authors: | Hou, Z, Su, L, Pei, J, Grishin, N.V, Zhang, H. | | Deposit date: | 2017-04-19 | | Release date: | 2017-06-07 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure of the CLOCK Transactivation Domain Exon19 in Complex with a Repressor.
Structure, 25, 2017
|
|
3CWI
 
 | | Crystal structure of thiamine biosynthesis protein (ThiS) from Geobacter metallireducens. Northeast Structural Genomics Consortium Target GmR137 | | Descriptor: | Thiamine-biosynthesis protein ThiS | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Mao, L, Janjua, H, Xiao, R, Maglaqui, M, Ciccosanti, C, Foote, E.L, Wang, H, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-04-21 | | Release date: | 2008-05-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of thiamine biosynthesis protein (ThiS) from Geobacter metallireducens.
To be Published
|
|
6I27
 
 | | Rea1 AAA2L-H2alpha deletion mutant in AMPPNP State | | Descriptor: | Midasin,Midasin,Midasin,Midasin,Midasin,Midasin,Midasin | | Authors: | Sosnowski, P, Urnavicius, L, Boland, A, Fagiewicz, R, Busselez, J, Papai, G, Schmidt, H. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | The CryoEM structure of the Saccharomyces cerevisiae ribosome maturation factor Rea1.
Elife, 7, 2018
|
|
1KN5
 
 | | SOLUTION STRUCTURE OF ARID DOMAIN OF ADR6 FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | Transcription regulatory protein ADR6 | | Authors: | Tu, X, Wu, J, Xu, Y, Shi, Y. | | Deposit date: | 2001-12-18 | | Release date: | 2002-07-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N resonance assignments and secondary structure of ADR6 DNA-binding domain.
J.Biomol.Nmr, 21, 2001
|
|
4BEV
 
 | | ATPase crystal structure with bound phosphate analogue | | Descriptor: | COPPER EFFLUX ATPASE, MAGNESIUM ION, TRIFLUOROMAGNESATE | | Authors: | Mattle, D, Drachmann, N.D, Liu, X.Y, Gourdon, P, Pedersen, B.P, Morth, P, Wang, J, Nissen, P. | | Deposit date: | 2013-03-12 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.583 Å) | | Cite: | ATPase Crystal Structure with Bound Phosphate Analogue
To be Published
|
|
3CTV
 
 | | Crystal structure of central domain of 3-hydroxyacyl-CoA dehydrogenase from Archaeoglobus fulgidus | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, SULFATE ION | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-04-14 | | Release date: | 2008-04-29 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | X-ray crystal structure of central domain of 3-hydroxyacyl-CoA dehydrogenase from Archaeoglobus fulgidus.
To be Published
|
|
5VNQ
 
 | | Neutron crystallographic structure of perdeuterated T4 lysozyme cysteine-free pseudo-wild type at cryogenic temperature | | Descriptor: | CHLORIDE ION, Endolysin | | Authors: | Li, L, Shukla, S, Meilleur, F, Standaert, R.F, Pierce, J, Myles, D.A.A, Cuneo, M.J. | | Deposit date: | 2017-05-01 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | NEUTRON DIFFRACTION (2.2 Å) | | Cite: | Neutron crystallographic studies of T4 lysozyme at cryogenic temperature.
Protein Sci., 26, 2017
|
|
6DTL
 
 | | Mitogen-activated protein kinase 6 | | Descriptor: | Mitogen-activated protein kinase 6 | | Authors: | Ruble, J. | | Deposit date: | 2018-06-17 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.753 Å) | | Cite: | Bipartite anchoring of SCREAM enforces stomatal initiation by coupling MAP kinases to SPEECHLESS.
Nat.Plants, 5, 2019
|
|
4B9W
 
 | | Structure of extended Tudor domain TD3 from mouse TDRD1 in complex with MILI peptide containing dimethylarginine 45. | | Descriptor: | GLYCEROL, PIWI-LIKE PROTEIN 2, TUDOR DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Mathioudakis, N, Palencia, A, Kadlec, J, Round, A, Tripsianes, K, Sattler, M, Pillai, R.S, Cusack, S. | | Deposit date: | 2012-09-08 | | Release date: | 2012-10-17 | | Last modified: | 2019-04-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The multiple Tudor domain-containing protein TDRD1 is a molecular scaffold for mouse Piwi proteins and piRNA biogenesis factors.
Rna, 18, 2012
|
|
4BEZ
 
 | | Night blindness causing G90D rhodopsin in the active conformation | | Descriptor: | ACETATE ION, PALMITIC ACID, RHODOPSIN, ... | | Authors: | Singhal, A, Ostermaier, M.K, Vishnivetskiy, S.A, Panneels, V, Homan, K.T, Tesmer, J.J.G, Veprintsev, D, Deupi, X, Gurevich, V.V, Schertler, G.F.X, Standfuss, J. | | Deposit date: | 2013-03-12 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Insights Into Congenital Stationary Night Blindness Based on the Structure of G90D Rhodopsin.
Embo Rep., 14, 2013
|
|
5VNE
 
 | |
4BGP
 
 | | Crystal structure of La Crosse virus nucleoprotein | | Descriptor: | GLYCEROL, NUCLEOPROTEIN, SULFATE ION | | Authors: | Reguera, J, Malet, H, Weber, F, Cusack, S. | | Deposit date: | 2013-03-28 | | Release date: | 2013-04-24 | | Last modified: | 2013-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Encapsidation of Genomic RNA by La Crosse Orthobunyavirus Nucleoprotein
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4B0T
 
 | | Structure of the Pup Ligase PafA of the Prokaryotic Ubiquitin-like Modification Pathway in Complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PUP--PROTEIN LIGASE | | Authors: | Ozcelik, D, Barandun, J, Schmitz, N, Sutter, M, Guth, E, Damberger, F.F, Allain, F.H.-T, Ban, N, Weber-Ban, E. | | Deposit date: | 2012-07-04 | | Release date: | 2012-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.159 Å) | | Cite: | Structures of Pup Ligase Pafa and Depupylase Dop from the Prokaryotic Ubiquitin-Like Modification Pathway.
Nat.Commun., 3, 2012
|
|
