2DCM
 
 | | The Crystal Structure of S603A Mutated Prolyl Tripeptidyl Aminopeptidase Complexed with Substrate | | Descriptor: | GLYCYLALANYL-N-2-NAPHTHYL-L-PROLINEAMIDE, dipeptidyl aminopeptidase IV, putative | | Authors: | Nakajima, Y, Ito, K, Xu, Y, Yamada, N, Onohara, Y, Yoshimoto, T. | | Deposit date: | 2006-01-09 | | Release date: | 2006-09-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure and Mechanism of Tripeptidyl Activity of Prolyl Tripeptidyl Aminopeptidase from Porphyromonas gingivalis
J.Mol.Biol., 362, 2006
|
|
6J3L
 
 | | Solution structure of the N-terminal extended protuberant domain of eukaryotic ribosomal stalk protein P0 | | Descriptor: | 60S acidic ribosomal protein P0 | | Authors: | Choi, K.H.A, Lee, K.M, Yang, L, Wing-Heng Yu, C, Banfield, D.K, Ito, K, Uchiumi, T, Wong, K.B. | | Deposit date: | 2019-01-04 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Mutagenesis Studies Evince the Role of the Extended Protuberant Domain of Ribosomal Protein uL10 in Protein Translation.
Biochemistry, 58, 2019
|
|
2DQM
 
 | | Crystal Structure of Aminopeptidase N complexed with bestatin | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, Aminopeptidase N, SULFATE ION, ... | | Authors: | Onohara, Y, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2006-05-29 | | Release date: | 2006-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Aminopeptidase N (proteobacteria alanyl aminopeptidase) from Escherichia coli: Crystal structure and conformational change of the methionine 260 residue involved in substrate recognition
J.Biol.Chem., 281, 2006
|
|
2DQ6
 
 | | Crystal Structure of Aminopeptidase N from Escherichia coli | | Descriptor: | Aminopeptidase N, SULFATE ION, ZINC ION | | Authors: | Nakajima, Y, Onohara, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2006-05-22 | | Release date: | 2006-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Aminopeptidase N (proteobacteria alanyl aminopeptidase) from Escherichia coli: Crystal structure and conformational change of the methionine 260 residue involved in substrate recognition
J.Biol.Chem., 281, 2006
|
|
2D5L
 
 | | Crystal Structure of Prolyl Tripeptidyl Aminopeptidase from Porphyromonas gingivalis | | Descriptor: | SULFATE ION, dipeptidyl aminopeptidase IV, putative | | Authors: | Nakajima, Y, Ito, K, Xu, Y, Yamada, N, Onohara, Y, Yoshimoto, T. | | Deposit date: | 2005-11-02 | | Release date: | 2006-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Mechanism of Tripeptidyl Activity of Prolyl Tripeptidyl Aminopeptidase from Porphyromonas gingivalis
J.Mol.Biol., 362, 2006
|
|
2Z3W
 
 | | Prolyl tripeptidyl aminopeptidase mutant E636A | | Descriptor: | Dipeptidyl aminopeptidase IV, GLYCEROL, SULFATE ION | | Authors: | Xu, Y, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2007-06-07 | | Release date: | 2008-02-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel inhibitor for prolyl tripeptidyl aminopeptidase from Porphyromonas gingivalis and details of substrate-recognition mechanism
J.Mol.Biol., 375, 2008
|
|
2Z3Z
 
 | | Prolyl tripeptidyl aminopeptidase mutant E636A complexd with an inhibitor | | Descriptor: | Dipeptidyl aminopeptidase IV, SULFATE ION, [(2R)-1-(L-ALANYL-L-ISOLEUCYL)PYRROLIDIN-2-YL]BORONIC ACID | | Authors: | Xu, Y, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2007-06-09 | | Release date: | 2008-02-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel inhibitor for prolyl tripeptidyl aminopeptidase from Porphyromonas gingivalis and details of substrate-recognition mechanism
J.Mol.Biol., 375, 2008
|
|
2ZXG
 
 | | Aminopeptidase N complexed with the aminophosphinic inhibitor of PL250, a transition state analogue | | Descriptor: | Aminopeptidase N, GLYCEROL, N-{(2S)-3-[(1R)-1-aminoethyl](hydroxy)phosphoryl-2-benzylpropanoyl}-L-phenylalanine, ... | | Authors: | Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2008-12-24 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of aminopeptidase N from Escherichia coli complexed with the transition-state analogue aminophosphinic inhibitor PL250
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2ZKY
 
 | | Crystal structure of human Cu-Zn superoxide dismutase mutant G93A | | Descriptor: | Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Yoshikawa, S, Kukimoto-Niino, M, Ito, K, Shirouzu, M, Urushitani, M, Takahashi, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-03-31 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human Cu-Zn superoxide dismutase mutant G93A
To be Published
|
|
2ZKX
 
 | | Crystal structure of human Cu-Zn superoxide dismutase mutant G85R in space group I212121 | | Descriptor: | COPPER (I) ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Yoshikawa, S, Kukimoto-Niino, M, Ito, K, Chen, L, Fu, Z.Q, Chrzas, J, Wang, B.C, Shirouzu, M, Urushitani, M, Takahashi, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-03-31 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structure of human Cu-Zn superoxide dismutase mutant G85R in space group I212121
To be Published
|
|
3AQP
 
 | | Crystal structure of SecDF, a translocon-associated membrane protein, from Thermus thrmophilus | | Descriptor: | Probable SecDF protein-export membrane protein | | Authors: | Tsukazaki, T, Mori, H, Echizen, Y, Ishitani, R, Fukai, S, Tanaka, T, Perederina, A, Vassylyev, D.G, Kohno, T, Ito, K, Nureki, O. | | Deposit date: | 2010-11-16 | | Release date: | 2011-05-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure and function of a membrane component SecDF that enhances protein export
Nature, 474, 2011
|
|
2ZJS
 
 | | Crystal Structure of SecYE translocon from Thermus thermophilus with a Fab fragment | | Descriptor: | Fab56 (heavy chain), Fab56 (light chain), Preprotein translocase SecE subunit, ... | | Authors: | Tsukazaki, T, Mori, H, Fukai, S, Ishitani, R, Perederina, A, Vassylyev, D.G, Ito, K, Nureki, O. | | Deposit date: | 2008-03-08 | | Release date: | 2008-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conformational transition of Sec machinery inferred from bacterial SecYE structures
Nature, 455, 2008
|
|
2ZKW
 
 | | Crystal structure of human Cu-Zn superoxide dismutase mutant G85R in space group P21 | | Descriptor: | COPPER (I) ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Yoshikawa, S, Kukimoto-Niino, M, Ito, K, Chen, L, Fu, Z.Q, Chrzas, J, Wang, B.C, Shirouzu, M, Urushitani, M, Takahashi, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-03-31 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human Cu-Zn superoxide dismutase mutant G85R in space group P21
To be Published
|
|
7DHW
 
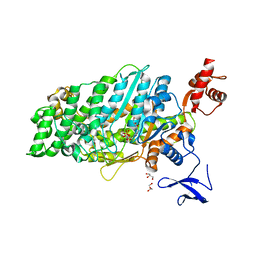 | | Crystal structure of myosin-XI motor domain in complex with ADP-ALF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Suzuki, K, Haraguchi, T, Tamanaha, M, Yoshimura, K, Imi, T, Tominaga, M, Sakayama, H, Nishiyama, T, Ito, K, Murata, T. | | Deposit date: | 2020-11-17 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery of ultrafast myosin, its amino acid sequence, and structural features.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2CZK
 
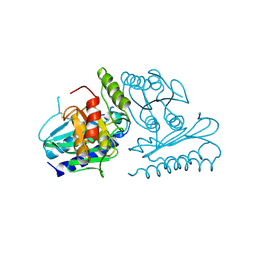 | | Crystal structure of human myo-inositol monophosphatase 2 (IMPA2) (trigonal form) | | Descriptor: | Inositol monophosphatase 2 | | Authors: | Arai, R, Ito, K, Hanawa-Suetsugu, K, Ohnishi, T, Ohba, H, Yoshikawa, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of human myo-inositol monophosphatase 2, the product of the putative susceptibility gene for bipolar disorder, schizophrenia, and febrile seizures
Proteins, 67, 2007
|
|
2CZI
 
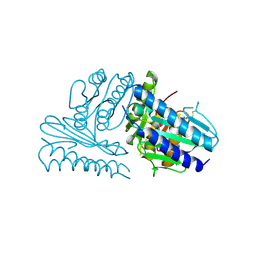 | | Crystal structure of human myo-inositol monophosphatase 2 (IMPA2) with calcium and phosphate ions | | Descriptor: | CALCIUM ION, Inositol monophosphatase 2, PHOSPHATE ION | | Authors: | Arai, R, Ito, K, Ohnishi, T, Ohba, H, Yoshikawa, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human myo-inositol monophosphatase 2, the product of the putative susceptibility gene for bipolar disorder, schizophrenia, and febrile seizures
Proteins, 67, 2007
|
|
2DDK
 
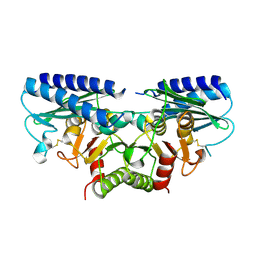 | | Crystal structure of human myo-inositol monophosphatase 2 (IMPA2) (orthorhombic form) | | Descriptor: | Inositol monophosphatase 2 | | Authors: | Arai, R, Ito, K, Kamo-Uchikubo, T, Bessho, Y, Ohba, H, Ohnishi, T, Yoshikawa, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-01-30 | | Release date: | 2007-02-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human myo-inositol monophosphatase 2, the product of the putative susceptibility gene for bipolar disorder, schizophrenia, and febrile seizures
Proteins, 67, 2007
|
|
2ECF
 
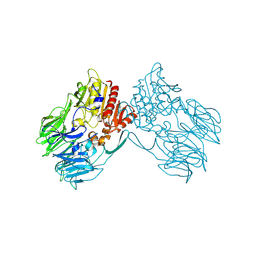 | |
6L37
 
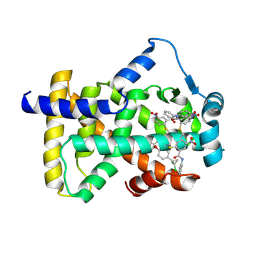 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-ciprofibrate co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-chloro-5-nitro-N-phenylbenzamide, 2-{4-[(1S)-2,2-dichlorocyclopropyl]phenoxy}-2-methylpropanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-10-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6L36
 
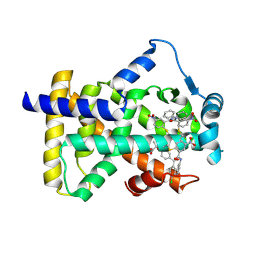 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-fenofibric acid co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-[4-(4-chlorobenzene-1-carbonyl)phenoxy]-2-methylpropanoic acid, 2-chloro-5-nitro-N-phenylbenzamide, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-10-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6L38
 
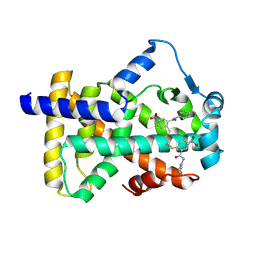 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-gemfibrozil co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-chloro-5-nitro-N-phenylbenzamide, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-10-09 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.761 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
5IWA
 
 | | Crystal structure of the 30S ribosomal subunit from Thermus thermophilus in complex with the GE81112 peptide antibiotic | | Descriptor: | (2S,3S)-2-{[(2S)-3-(2-amino-1H-imidazol-5-yl)-2-{[(2S,4S)-5-(carbamoyloxy)-4-hydroxy-2-({[(2S,3S)-3-hydroxypiperidin-2-yl]carbonyl}amino)pentanoyl]amino}propanoyl]amino}-3-(2-chloro-1H-imidazol-5-yl)-3-hydroxypropanoic acid, 16S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Schedlbauer, A, Kaminishi, T, Ochoa-Lizarralde, B, Chieko, N, Masahito, K, Takemoto, C, Yokoyama, S, Connell, S.R, Fucini, P. | | Deposit date: | 2016-03-22 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Inhibition of translation initiation complex formation by GE81112 unravels a 16S rRNA structural switch involved in P-site decoding.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7CSL
 
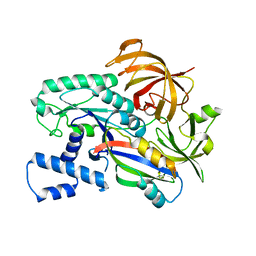 | | Crystal structure of the archaeal EF1A-EF1B complex | | Descriptor: | Elongation factor 1-alpha, Elongation factor 1-beta | | Authors: | Suzuki, T, Ito, K, Miyoshi, T, Murakami, R, Uchiumi, T. | | Deposit date: | 2020-08-15 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the Switching Off of the Interaction between the Archaeal Ribosomal Stalk and aEF1A by Nucleotide Exchange Factor aEF1B.
J.Mol.Biol., 433, 2021
|
|
6LX4
 
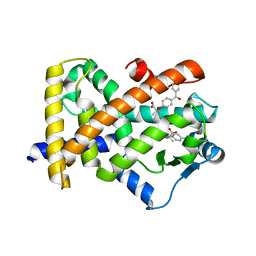 | | X-ray structure of human PPARalpha ligand binding domain-fenofibric acid co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-[4-(4-chlorobenzene-1-carbonyl)phenoxy]-2-methylpropanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LX9
 
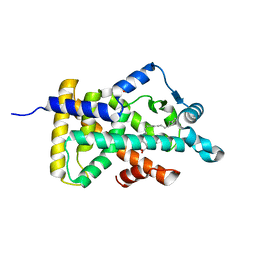 | | X-ray structure of human PPARalpha ligand binding domain-arachidonic acid co-crystals obtained by delipidation and cross-seeding | | Descriptor: | ARACHIDONIC ACID, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
