2WBM
 
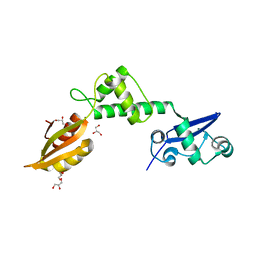 | | Crystal structure of mthSBDS, the homologue of the Shwachman-Bodian- Diamond syndrome protein in the euriarchaeon Methanothermobacter thermautotrophicus | | Descriptor: | CHLORIDE ION, GLYCEROL, RIBOSOME MATURATION PROTEIN SDO1 HOMOLOG, ... | | Authors: | Ng, C.L, Isupov, M.N, Lebedev, A.A, Ortiz-Lombardia, M, Antson, A.A. | | Deposit date: | 2009-03-02 | | Release date: | 2009-06-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Conformational Flexibility and Molecular Interactions of an Archaeal Homologue of the Shwachman-Bodian-Diamond Syndrome Protein.
Bmc Struct.Biol., 9, 2009
|
|
4C3T
 
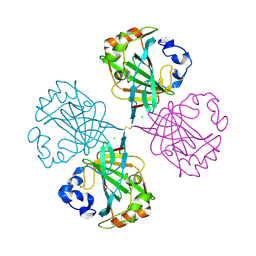 | | The Carbonic anhydrase from Thermovibrio ammonificans reveals an interesting intermolecular disulfide contributing to increasing thermal stability of this enzyme | | Descriptor: | CARBONATE DEHYDRATASE, CHLORIDE ION, ZINC ION | | Authors: | James, P, Isupov, M, Sayer, C, Berg, S, Lioliou, M, Kotlar, H, Littlechild, J. | | Deposit date: | 2013-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The Structure of a Tetrameric [Alpha]-Carbonic Anhydrase from Thermovibrio Ammonificans Reveals a Core Formed Around Intermolecular Disulfides that Contribute to its Thermostability
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4C6H
 
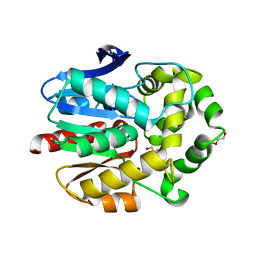 | | Haloalkane dehalogenase with 1-hexanol | | Descriptor: | CHLORIDE ION, HALOALKANE DEHALOGENASE, HEXAN-1-OL, ... | | Authors: | Novak, H.R, Sayer, C, Isupov, M, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Biochemical and Structural Characterisation of a Haloalkane Dehalogenase from a Marine Rhodobacteraceae.
FEBS Lett., 588, 2014
|
|
4BRZ
 
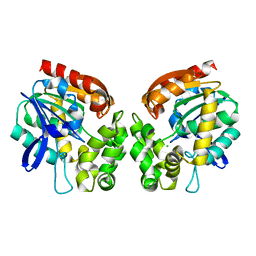 | | Haloalkane dehalogenase | | Descriptor: | CHLORIDE ION, HALOALKANE DEHALOGENASE | | Authors: | Novak, H.R, Sayer, C, Isupov, M, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2013-06-06 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Biochemical and Structural Characterisation of a Haloalkane Dehalogenase from a Marine Rhodobacteraceae.
FEBS Lett., 588, 2014
|
|
2WB9
 
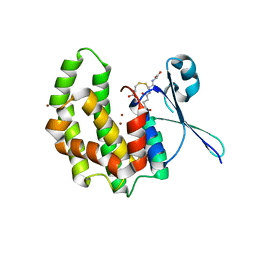 | | Fasciola hepatica sigma class GST | | Descriptor: | BROMIDE ION, CYSTEINE, GLUTATHIONE, ... | | Authors: | Line, K, Isupov, M.N, LaCourse, J, Brophy, P.M, Littlechild, J.A. | | Deposit date: | 2009-02-23 | | Release date: | 2010-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The 1.6 Angstrom Crystal Structure of the Fasciola Hepatica Sigma Class Gst
To be Published
|
|
2YN4
 
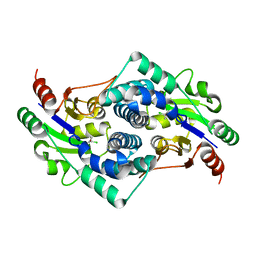 | | L-2-chlorobutryic acid bound complex L-haloacid dehalogenase from a Rhodobacteraceae family bacterium | | Descriptor: | (2S)-2-chlorobutanoic acid, L-HALOACID DEHALOGENASE | | Authors: | Novak, H.R, Sayer, C, Isupov, M.N, Paszkiewicz, K, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2012-10-12 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Marine Rhodobacteraceae L-Haloacid Dehalogenase Contains a Novel His/Glu Dyad that Could Activate the Catalytic Water.
FEBS J., 280, 2013
|
|
2W43
 
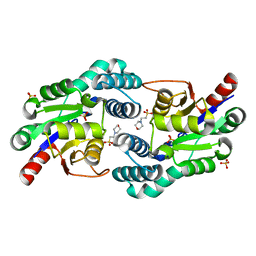 | | Structure of L-haloacid dehalogenase from S. tokodaii | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HYPOTHETICAL 2-HALOALKANOIC ACID DEHALOGENASE, PHOSPHATE ION | | Authors: | Rye, C.A, Isupov, M.N, Lebedev, A.A, Littlechild, J.A. | | Deposit date: | 2008-11-21 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Biochemical and Structural Studies of a L-Haloacid Dehalogenase from the Thermophilic Archaeon Sulfolobus Tokodaii.
Extremophiles, 13, 2009
|
|
2WS2
 
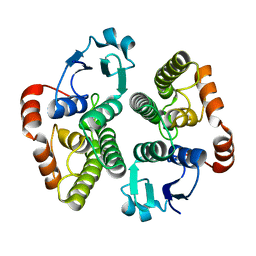 | |
2YMM
 
 | | Sulfate bound L-haloacid dehalogenase from a Rhodobacteraceae family bacterium | | Descriptor: | L-HALOACID DEHALOGENASE, SULFATE ION | | Authors: | Novak, H.R, Sayer, C, Isupov, M.N, Paszkiewicz, K, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2012-10-09 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Marine Rhodobacteraceae L-Haloacid Dehalogenase Contains a Novel His/Glu Dyad that Could Activate the Catalytic Water.
FEBS J., 280, 2013
|
|
2YMP
 
 | | Chloroacetic acid complex bound L-haloacid dehalogenase from a Rhodobacteraceae family bacterium | | Descriptor: | L-HALOACID DEHALOGENASE | | Authors: | Novak, H.R, Sayer, C, Isupov, M.N, Paszkiewicz, K, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2012-10-10 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Marine Rhodobacteraceae L-Haloacid Dehalogenase Contains a Novel His/Glu Dyad that Could Activate the Catalytic Water.
FEBS J., 280, 2013
|
|
2YMQ
 
 | | Chloropropionic acid complex bound L-haloacid dehalogenase from a Rhodobacteraceae family bacterium | | Descriptor: | L-HALOACID DEHALOGENASE | | Authors: | Novak, H.R, Sayer, C, Isupov, M.N, Paszkiewicz, K, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2012-10-10 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Marine Rhodobacteraceae L-Haloacid Dehalogenase Contains a Novel His/Glu Dyad that Could Activate the Catalytic Water.
FEBS J., 280, 2013
|
|
2YML
 
 | | Native L-haloacid dehalogenase from a Rhodobacteraceae family bacterium | | Descriptor: | L-HALOACID DEHALOGENASE | | Authors: | Novak, H.R, Sayer, C, Isupov, M.N, Paszkiewicz, K, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2012-10-09 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Marine Rhodobacteraceae L-Haloacid Dehalogenase Contains a Novel His/Glu Dyad that Could Activate the Catalytic Water.
FEBS J., 280, 2013
|
|
4CMD
 
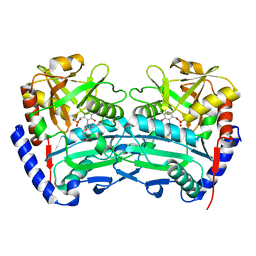 | | The (R)-selective transaminase from Nectria haematococca | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sayer, C, Isupov, M, Martinez-Torres, R.J, Richter, N, Hailes, H.C, Ward, J, Littlechild, J. | | Deposit date: | 2014-01-16 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The Substrate Specificity, Enantioselectivity and Structure of the (R)-Selective Amine:Pyruvate Transaminase from Nectria Haematococca.
FEBS J., 281, 2014
|
|
3ZRR
 
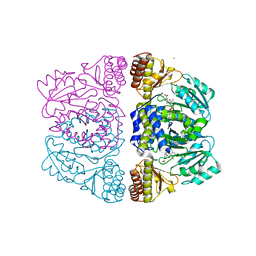 | | Crystal structure and substrate specificity of a thermophilic archaeal serine : pyruvate aminotransferase from Sulfolobus solfataricus | | Descriptor: | 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, CALCIUM ION, SERINE-PYRUVATE AMINOTRANSFERASE (AGXT) | | Authors: | Sayer, C, Bommer, M, Isupov, M.N, Ward, J, Littlechild, J. | | Deposit date: | 2011-06-17 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure and Substrate Specificity of the Thermophilic Serine:Pyruvate Aminotransferase from Sulfolobus Solfataricus
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4BQN
 
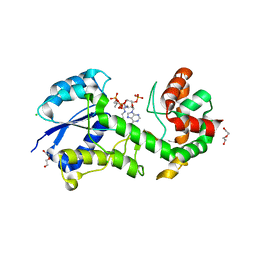 | | Structural insights into WcbI, a novel polysaccharide biosynthesis enzyme. Native protein. | | Descriptor: | CAPSULAR POLYSACCHARIDE BIOSYNTHESIS PROTEIN, CHLORIDE ION, COENZYME A, ... | | Authors: | Vivoli, M, Ayres, E, Isupov, M.N, Harmer, N.J. | | Deposit date: | 2013-05-31 | | Release date: | 2013-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural Insights Into Wcbi, a Novel Polysaccharide-Biosynthesis Enzyme.
Iucrj, 1, 2014
|
|
4BQO
 
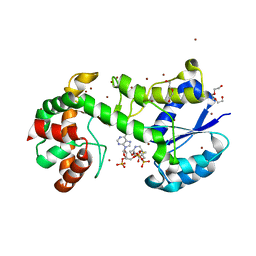 | | Structural insights into WcbI, a novel polysaccharide biosynthesis enzyme. Native protein without disulfide bond between COA and Cys14. | | Descriptor: | BROMIDE ION, COENZYME A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vivoli, M, Ayres, E, Isupov, M.N, Harmer, N.J. | | Deposit date: | 2013-05-31 | | Release date: | 2013-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural Insights Into Wcbi, a Novel Polysaccharide-Biosynthesis Enzyme.
Iucrj, 1, 2014
|
|
3ZRP
 
 | | Crystal structure and substrate specificity of a thermophilic archaeal serine : pyruvate aminotransferase from Sulfolobus solfataricus | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SERINE-PYRUVATE AMINOTRANSFERASE (AGXT) | | Authors: | Sayer, C, Bommer, M, Isupov, M.N, Ward, J, Littlechild, J. | | Deposit date: | 2011-06-17 | | Release date: | 2012-06-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure and Substrate Specificity of the Thermophilic Serine:Pyruvate Aminotransferase from Sulfolobus Solfataricus
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3ZRQ
 
 | | Crystal structure and substrate specificity of a thermophilic archaeal serine : pyruvate aminotransferase from Sulfolobus solfataricus | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, SERINE-PYRUVATE AMINOTRANSFERASE (AGXT) | | Authors: | Sayer, C, Bommer, M, Isupov, M.N, Ward, J, Littlechild, J. | | Deposit date: | 2011-06-17 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Substrate Specificity of the Thermophilic Serine:Pyruvate Aminotransferase from Sulfolobus Solfataricus
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4B9B
 
 | | The structure of the omega aminotransferase from Pseudomonas aeruginosa | | Descriptor: | BETA-ALANINE-PYRUVATE TRANSAMINASE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sayer, C, Isupov, M.N, Westlake, A, Littlechild, J.A. | | Deposit date: | 2012-09-03 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Studies with Pseudomonas and Chromobacterium [Omega]-Aminotransferases Provide Insights Into Their Differing Substrate Specificity.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4B98
 
 | | The structure of the omega aminotransferase from Pseudomonas aeruginosa | | Descriptor: | 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, BETA-ALANINE--PYRUVATE TRANSAMINASE, CALCIUM ION, ... | | Authors: | Sayer, C, Isupov, M.N, Westlake, A, Littlechild, J.A. | | Deposit date: | 2012-09-03 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Studies with Pseudomonas and Chromobacterium [Omega]-Aminotransferases Provide Insights Into Their Differing Substrate Specificity.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BA4
 
 | |
4AH3
 
 | |
4BA5
 
 | | Crystal structure of omega-transaminase from Chromobacterium violaceum | | Descriptor: | 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, AMINOTRANSFERASE, SULFATE ION | | Authors: | Sayer, C, Isupov, M.N, Littlechild, J.A. | | Deposit date: | 2012-09-11 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Studies with Pseudomonas and Chromobacterium [Omega]-Aminotransferases Provide Insights Into Their Differing Substrate Specificity.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4C76
 
 | |
4CF3
 
 | |
