4HAI
 
 | | Crystal structure of human soluble epoxide hydrolase complexed with N-cycloheptyl-1-(mesitylsulfonyl)piperidine-4-carboxamide. | | Descriptor: | Bifunctional epoxide hydrolase 2, MAGNESIUM ION, N-cycloheptyl-1-[(2,4,6-trimethylphenyl)sulfonyl]piperidine-4-carboxamide, ... | | Authors: | Pecic, S, Pakhomova, S, Newcomer, M.E, Morisseau, C, Hammock, B.D, Zhu, Z, Deng, S. | | Deposit date: | 2012-09-26 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Synthesis and structure-activity relationship of piperidine-derived non-urea soluble epoxide hydrolase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1S8O
 
 | | Human soluble Epoxide Hydrolase | | Descriptor: | HEXAETHYLENE GLYCOL, epoxide hydrolase 2, cytoplasmic | | Authors: | Gomez, G.A, Morisseau, C, Hammock, B.D, Christianson, D.W. | | Deposit date: | 2004-02-03 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human epoxide hydrolase reveals mechanistic inferences on bifunctional catalysis in epoxide and phosphate ester hydrolysis
Biochemistry, 43, 2004
|
|
3E51
 
 | | Crystal structure of HCV NS5B polymerase with a novel pyridazinone inhibitor | | Descriptor: | N-{3-[5-hydroxy-2-(3-methylbutyl)-3-oxo-6-pyrrolidin-1-yl-2,3-dihydropyridazin-4-yl]-1,1-dioxido-2H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Han, Q, Showalter, R.E, Zhao, Q, Kissinger, C.R. | | Deposit date: | 2008-08-12 | | Release date: | 2009-08-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel HCV NS5B polymerase inhibitors derived from 4-(1',1'-dioxo-1',4'-dihydro-1'lambda(6)-benzo[1',2',4']thiadiazin-3'-yl)-5-hydroxy-2H-pyridazin-3-ones. Part 5: Exploration of pyridazinones containing 6-amino-substituents.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1ENH
 
 | | STRUCTURAL STUDIES OF THE ENGRAILED HOMEODOMAIN | | Descriptor: | ENGRAILED HOMEODOMAIN | | Authors: | Clarke, N.D, Kissinger, C.R, Desjarlais, J, Gilliland, G.L, Pabo, C.O. | | Deposit date: | 1994-05-20 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural studies of the engrailed homeodomain.
Protein Sci., 3, 1994
|
|
1ZD4
 
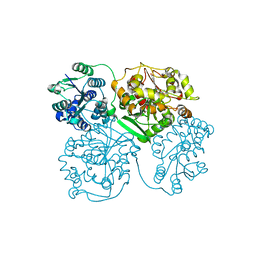 | | Human soluble epoxide hydrolase 4-(3-cyclohexyluriedo)-hexanoic acid complex | | Descriptor: | 6-{[(CYCLOHEXYLAMINO)CARBONYL]AMINO}HEXANOIC ACID, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Gomez, G.A, Morisseau, C, Hammock, B.D, Christianson, D.W. | | Deposit date: | 2005-04-14 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human soluble epoxide hydrolase: structural basis of inhibition by 4-(3-cyclohexylureido)-carboxylic acids
Protein Sci., 15, 2006
|
|
1ZD3
 
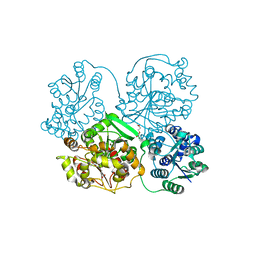 | | Human soluble epoxide hydrolase 4-(3-cyclohexyluriedo)-butyric acid complex | | Descriptor: | 4-{[(CYCLOHEXYLAMINO)CARBONYL]AMINO}BUTANOIC ACID, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Gomez, G.A, Morisseau, C, Hammock, B.D, Christianson, D.W. | | Deposit date: | 2005-04-14 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human soluble epoxide hydrolase: structural basis of inhibition by 4-(3-cyclohexylureido)-carboxylic acids
Protein Sci., 15, 2006
|
|
1ZD5
 
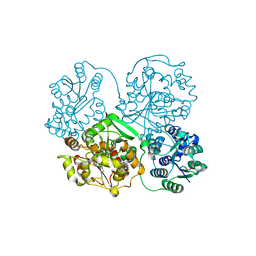 | | Human soluble epoxide hydrolase 4-(3-cyclohexyluriedo)-heptanoic acid complex | | Descriptor: | 7-{[(CYCLOHEXYLAMINO)CARBONYL]AMINO}HEPTANOIC ACID, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Gomez, G.A, Morisseau, C, Hammock, B.D, Christianson, D.W. | | Deposit date: | 2005-04-14 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human soluble epoxide hydrolase: structural basis of inhibition by 4-(3-cyclohexylureido)-carboxylic acids
Protein Sci., 15, 2006
|
|
1ZD2
 
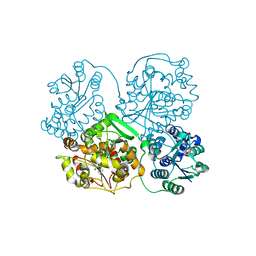 | | Human soluble epoxide hydrolase 4-(3-cyclohexyluriedo)-ethanoic acid complex | | Descriptor: | MAGNESIUM ION, N-[(CYCLOHEXYLAMINO)CARBONYL]GLYCINE, PHOSPHATE ION, ... | | Authors: | Gomez, G.A, Morisseau, C, Hammock, B.D, Christianson, D.W. | | Deposit date: | 2005-04-14 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human soluble epoxide hydrolase: structural basis of inhibition by 4-(3-cyclohexylureido)-carboxylic acids
Protein Sci., 15, 2006
|
|
3CWJ
 
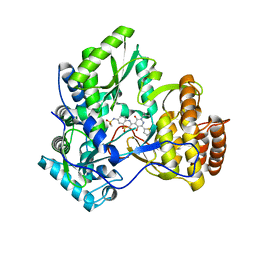 | | Crystal structure of hcv ns5b polymerase with a novel pyridazinone inhibitor | | Descriptor: | N-{3-[5-hydroxy-2-(3-methylbutyl)-3-oxo-6-thiophen-2-yl-2,3-dihydropyridazin-4-yl]-1,1-dioxido-2H-1,4-benzothiazin-7-yl}methanesulfonamide, RNA-DIRECTED RNA POLYMERASE | | Authors: | Han, Q, Showalter, R.E, Zhao, Q, Kissinger, C.R. | | Deposit date: | 2008-04-21 | | Release date: | 2009-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 4-(1,1-Dioxo-1,4-dihydro-1lambda6-benzo[1,4]thiazin-3-yl)-5-hydroxy-2H-pyridazin-3-ones as potent inhibitors of HCV NS5B polymerase
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3CO9
 
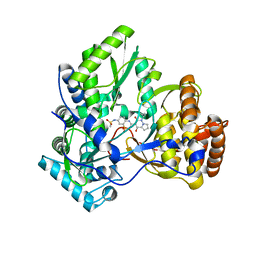 | | Crystal structure of HCV NS5B polymerase with a novel pyridazinone inhibitor | | Descriptor: | N-{3-[4-hydroxy-1-(3-methylbutyl)-2-oxo-1,2-dihydropyrrolo[1,2-b]pyridazin-3-yl]-1,1-dioxido-2H-1,2,4-benzothiadiazin-7 -yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Han, Q, Showalter, R.E, Zhao, Q, Kissinger, C.R. | | Deposit date: | 2008-03-27 | | Release date: | 2009-02-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyrrolo[1,2-b]pyridazin-2-ones as potent inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3GYN
 
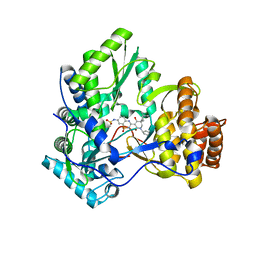 | | Crystal structure of HCV NS5B polymerase with a novel monocyclic dihydropyridinone inhibitor | | Descriptor: | N-{3-[(5R)-1-cyclopentyl-4-hydroxy-5-methyl-5-(3-methylbutyl)-2-oxo-1,2,5,6-tetrahydropyridin-3-yl]-1,1-dioxido-4H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2009-04-04 | | Release date: | 2009-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 5,5'- and 6,6'-dialkyl-5,6-dihydro-1H-pyridin-2-ones as potent inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3IGV
 
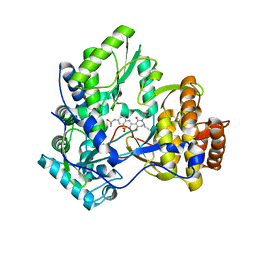 | | Crystal structure of HCV NS5B polymerase with a novel monocyclic dihydro-pyridinone inhibitor | | Descriptor: | N-{3-[(6S)-6-ethyl-1-(4-fluorobenzyl)-4-hydroxy-2-oxo-1,2,5,6-tetrahydropyridin-3-yl]-1,1-dioxido-2H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-DIRECTED RNA POLYMERASE | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2009-07-28 | | Release date: | 2009-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 5,5'- and 6,6'-dialkyl-5,6-dihydro-1H-pyridin-2-ones as potent inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1CR6
 
 | | CRYSTAL STRUCTURE OF MURINE SOLUBLE EPOXIDE HYDROLASE COMPLEXED WITH CPU INHIBITOR | | Descriptor: | EPOXIDE HYDROLASE, N-CYCLOHEXYL-N'-(PROPYL)PHENYL UREA | | Authors: | Argiriadi, M.A, Morisseau, C, Hammock, B.D, Christianson, D.W. | | Deposit date: | 1999-08-13 | | Release date: | 1999-11-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Detoxification of environmental mutagens and carcinogens: structure, mechanism, and evolution of liver epoxide hydrolase.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3CVK
 
 | | Crystal structure of HCV NS5B polymerase with a novel pyridazinone inhibitor | | Descriptor: | N-{3-[1-(3,3-Dimethyl-butyl)-4-hydroxy-2-oxo-1,2,4a,5,6,7-hexahydro-pyrrolo[1,2-b]pyridazin-3-yl]-1,1-dioxo-1,2-dihydro -1lambda6-benzo[1,2,4]thiadiazin-7-yl}-methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2008-04-18 | | Release date: | 2009-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Hexahydro-pyrrolo- and hexahydro-1H-pyrido[1,2-b]pyridazin-2-ones as potent inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6CK5
 
 | | PRPP riboswitch from T. mathranii bound to PRPP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, BARIUM ION, MAGNESIUM ION, ... | | Authors: | Knappenberger, A.J, Reiss, C.W, Strobel, S.A. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structures of two aptamers with differing ligand specificity reveal ruggedness in the functional landscape of RNA.
Elife, 7, 2018
|
|
1EK1
 
 | | CRYSTAL STRUCTURE OF MURINE SOLUBLE EPOXIDE HYDROLASE COMPLEXED WITH CIU INHIBITOR | | Descriptor: | EPOXIDE HYDROLASE, N-CYCLOHEXYL-N'-(4-IODOPHENYL)UREA | | Authors: | Argiriadi, M.A, Morisseau, C, Goodrow, M.H, Dowdy, D.L, Hammock, B.D, Christianson, D.W. | | Deposit date: | 2000-03-06 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Binding of alkylurea inhibitors to epoxide hydrolase implicates active site tyrosines in substrate activation.
J.Biol.Chem., 275, 2000
|
|
1EK2
 
 | | CRYSTAL STRUCTURE OF MURINE SOLUBLE EPOXIDE HYDROLASE COMPLEXED WITH CDU INHIBITOR | | Descriptor: | EPOXIDE HYDROLASE, N-CYCLOHEXYL-N'-DECYLUREA | | Authors: | Argiriadi, M.A, Morisseau, C, Goodrow, M.H, Dowdy, D.L, Hammock, B.D, Christianson, D.W. | | Deposit date: | 2000-03-06 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Binding of alkylurea inhibitors to epoxide hydrolase implicates active site tyrosines in substrate activation.
J.Biol.Chem., 275, 2000
|
|
1TE1
 
 | | Crystal structure of family 11 xylanase in complex with inhibitor (XIP-I) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, endo-1,4-xylanase, ... | | Authors: | Payan, F, Leone, P, Furniss, C, Tahir, T, Durand, A, Porciero, S, Manzanares, P, Williamson, G, Gilbert, H.J, Juge, N, Roussel, A. | | Deposit date: | 2004-05-24 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Dual Nature of the Wheat Xylanase Protein Inhibitor XIP-I: STRUCTURAL BASIS FOR THE INHIBITION OF FAMILY 10 AND FAMILY 11 XYLANASES.
J.Biol.Chem., 279, 2004
|
|
6FWH
 
 | | Acanthamoeba IGPD in complex with R-C348 to 1.7A resolution | | Descriptor: | Imidazoleglycerol-phosphate dehydratase, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Roberts, C.W, Bisson, C, Baker, P.J. | | Deposit date: | 2018-03-06 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural and functional studies of histidine biosynthesis in Acanthamoeba spp. demonstrates a novel molecular arrangement and target for antimicrobials.
PLoS ONE, 13, 2018
|
|
1TA3
 
 | | Crystal Structure of xylanase (GH10) in complex with inhibitor (XIP) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Endo-1,4-beta-xylanase, ... | | Authors: | Payan, F, Leone, P, Furniss, C, Tahir, T, Durand, A, Porciero, S, Manzanares, P, Williamson, G, Gilbert, H.J, Juge, N, Roussel, A. | | Deposit date: | 2004-05-19 | | Release date: | 2004-07-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Dual Nature of the Wheat Xylanase Protein Inhibitor XIP-I: STRUCTURAL BASIS FOR THE INHIBITION OF FAMILY 10 AND FAMILY 11 XYLANASES.
J.Biol.Chem., 279, 2004
|
|
6AUM
 
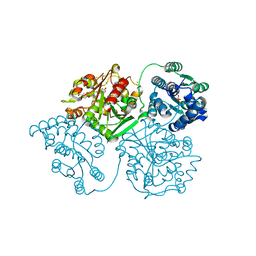 | | Crystal structure of human soluble epoxide hydrolase complexed with trans-4-[4-(3-trifluoromethoxyphenyl-l-ureido)-cyclohexyloxy]-benzoic acid. | | Descriptor: | 4-{[trans-4-({[4-(trifluoromethoxy)phenyl]carbamoyl}amino)cyclohexyl]oxy}benzoic acid, Bifunctional epoxide hydrolase 2, CHLORIDE ION, ... | | Authors: | Kodani, S.D, Bahkta, S, Hwang, S.H, Pakhomova, S, Newcomer, M.E, Morisseau, C, Hammock, B. | | Deposit date: | 2017-09-01 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Identification and optimization of soluble epoxide hydrolase inhibitors with dual potency towards fatty acid amide hydrolase.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6GH0
 
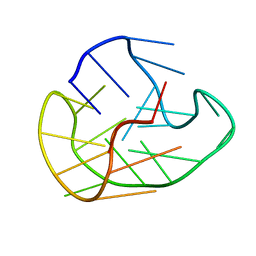 | |
6GQ3
 
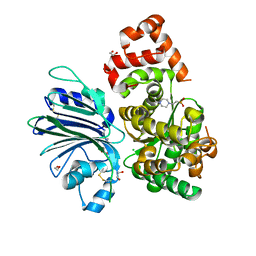 | | Human asparagine synthetase (ASNS) in complex with 6-diazo-5-oxo-L-norleucine (DON) at 1.85 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-OXO-L-NORLEUCINE, ... | | Authors: | Zhu, W, Radadiya, A, Bisson, C, Jin, Y, Nordin, B.E, Imasaki, T, Wenzel, S, Sedelnikova, S.E, Berry, A.H, Nomanbhoy, T.K, Kozarich, J.W, Takagi, Y, Rice, D.W, Richards, N.G.J. | | Deposit date: | 2018-06-07 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | High-resolution crystal structure of human asparagine synthetase enables analysis of inhibitor binding and selectivity.
Commun Biol, 2, 2019
|
|
6HDI
 
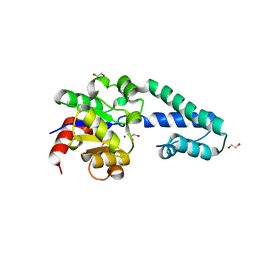 | |
6HDL
 
 | | R49K variant of beta-phosphoglucomutase from Lactococcus lactis complexed with magnesium trifluoride and beta-G6P to 1.2 A. | | Descriptor: | 1,2-ETHANEDIOL, 6-O-phosphono-beta-D-glucopyranose, Beta-phosphoglucomutase, ... | | Authors: | Wood, H.P, Robertson, A.J, Bisson, C, Waltho, J.P. | | Deposit date: | 2018-08-17 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Transition state of phospho-enzyme hydrolysis in beta-phosphoglucomutase.
To Be Published
|
|
