3UY8
 
 | | Designed protein KE59 R5_11/5F | | Descriptor: | Kemp eliminase KE59 R5_11/5F, SULFATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-06 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3RON
 
 | | Crystal Structure and Hemolytic Activity of the Cyt1Aa Toxin from Bacillus thuringiensis subsp. israelensis | | Descriptor: | Type-1Aa cytolytic delta-endotoxin | | Authors: | Cohen, S, Albeck, S, Ben-Dov, E, Cahan, R, Firer, M, Zaritsky, A, Dym, O, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-04-26 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Cyt1Aa Toxin: Crystal Structure Reveals Implications for Its Membrane-Perforating Function.
J.Mol.Biol., 413, 2011
|
|
3UXD
 
 | | Designed protein KE59 R1 7/10H with dichlorobenzotriazole (DBT) | | Descriptor: | 5,7-dichloro-1H-benzotriazole, Kemp eliminase KE59 R1 7/10H, PHOSPHATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-05 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UY7
 
 | | Designed protein KE59 R1 7/10H with G130S mutation | | Descriptor: | Kemp eliminase KE59 R1 7/10H, SODIUM ION, SULFATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-06 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UXA
 
 | | Designed protein KE59 R1 7/10H | | Descriptor: | Kemp eliminase KE59 R1 7/10H, PHOSPHATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-05 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UZ5
 
 | | Designed protein KE59 R13 3/11H | | Descriptor: | 5,7-dichloro-1H-benzotriazole, Kemp eliminase KE59 R13 3/11H, PHOSPHATE ION, ... | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-07 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UYC
 
 | | Designed protein KE59 R8_2/7A | | Descriptor: | Kemp eliminase KE59 R8_2/7A, PHOSPHATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-06 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3NPU
 
 | | Optimization of the in silico designed Kemp eliminase KE70 by computational design and directed evolution | | Descriptor: | deoxyribose phosphate aldolase | | Authors: | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Dym, O, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2010-06-29 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
3NPW
 
 | | In silico designed of an improved Kemp eliminase KE70 mutant by computational design and directed evolution | | Descriptor: | deoxyribose phosphate aldolase | | Authors: | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Dym, O, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2010-06-29 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
3NQV
 
 | | Optimization of the in silico designed Kemp eliminase KE70 by computational design and directed evolution R5 7/4A | | Descriptor: | BENZAMIDINE, deoxyribose phosphate aldolase | | Authors: | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Dym, O, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2010-06-29 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
3Q9U
 
 | | In silico and in vitro co-evolution of a high affinity complementary protein-protein interface | | Descriptor: | COENZYME A, CoA binding protein, consensus ankyrin repeat | | Authors: | Karanicolas, J, Corn, J.E, Chen, I, Joachimiak, L.A, Dym, O, Chung, S, Albeck, S, Unger, T, Hu, W, Liu, G, Delbecq, S, Montelione, G.T, Spiegel, C, Liu, D, Baker, D, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-01-10 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A de novo protein binding pair by computational design and directed evolution.
Mol.Cell, 42, 2011
|
|
3Q2D
 
 | | Optimization of the in silico designed Kemp eliminase KE70 by computational design and directed evolution | | Descriptor: | 5-nitro-1H-benzotriazole, Deoxyribose phosphate aldolase | | Authors: | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Murphy, P, Dym, O, Albeck, S, Kiss, G, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2010-12-20 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
3Q9N
 
 | | In silico and in vitro co-evolution of a high affinity complementary protein-protein interface | | Descriptor: | CARBAMOYL SARCOSINE, COENZYME A, CoA binding protein, ... | | Authors: | Karanicolas, J, Corn, J.E, Chen, I, Joachimiak, L.A, Dym, O, Chung, S, Albeck, S, Unger, T, Hu, W, Liu, G, Delbecq, S, Montelione, G.T, Spiegel, C, Liu, D, Baker, D, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-01-09 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A de novo protein binding pair by computational design and directed evolution.
Mol.Cell, 42, 2011
|
|
3UTZ
 
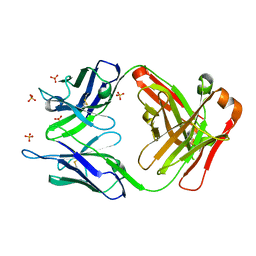 | | Endogenous-like inhibitory antibodies targeting activated metalloproteinase motifs show therapeutic potential | | Descriptor: | Metalloproteinase, heavy chain, light chain, ... | | Authors: | Sela-Passwell, N, Kikkeri, R, Dym, O, Rozenberg, H, Margalit, R, Arad-Yellin, R, Eisenstein, M, Brenner, O, Shoham, T, Danon, T, Shanzer, A, Sagi, I, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-11-27 | | Release date: | 2011-12-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Antibodies targeting the catalytic zinc complex of activated matrix metalloproteinases show therapeutic potential.
NAT.MED. (N.Y.), 18, 2012
|
|
1XXM
 
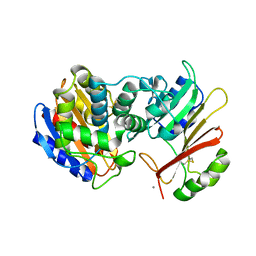 | | The modular architecture of protein-protein binding site | | Descriptor: | Beta-lactamase TEM, Beta-lactamase inhibitory protein, CALCIUM ION | | Authors: | Reichmann, D, Rahat, O, Albeck, S, Meged, R, Dym, O, Schreiber, G, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2004-11-07 | | Release date: | 2005-01-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The modular architecture of protein-protein binding interfaces
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1Y7V
 
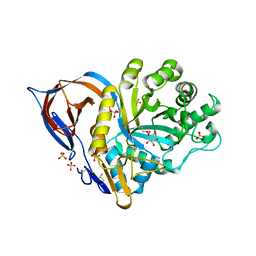 | | X-ray structure of human acid-beta-glucosidase covalently bound to conduritol B epoxide | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glucosylceramidase, ... | | Authors: | Premkumar, L, Sawkar, A.R, Boldin-Adamsky, S, Toker, L, Silman, I, Kelly, J.W, Futerman, A.H, Sussman, J.L, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2004-12-10 | | Release date: | 2005-04-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structure of human acid-beta-glucosidase covalently bound to conduritol-B-epoxide. Implications for Gaucher disease.
J.Biol.Chem., 280, 2005
|
|
1ZGC
 
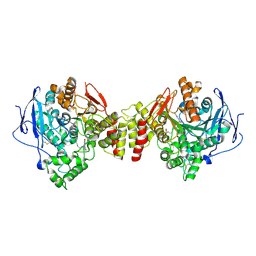 | | Crystal Structure of Torpedo Californica Acetylcholinesterase in Complex With an (RS)-Tacrine(10)-Hupyridone Inhibitor. | | Descriptor: | (5S)-5-{[10-(1,2,3,4-TETRAHYDROACRIDIN-9-YLAMINO)DECYL]AMINO}-5,6,7,8-TETRAHYDROQUINOLIN-2(1H)-ONE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Haviv, H, Wong, D.M, Greenblatt, H.M, Carlier, P.R, Pang, Y.P, Silman, I, Sussman, J.L, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2005-04-21 | | Release date: | 2005-08-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Packing Mediates Enantioselective Ligand Recognition at the Peripheral Site of Acetylcholinesterase
J.Am.Chem.Soc., 127, 2005
|
|
1Y7W
 
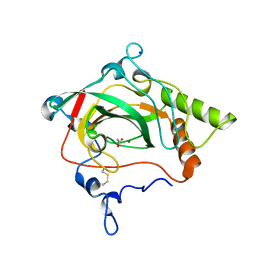 | | Crystal structure of a halotolerant carbonic anhydrase from Dunaliella salina | | Descriptor: | ACETIC ACID, Halotolerant alpha-type carbonic anhydrase (dCA II), SODIUM ION, ... | | Authors: | Premkumar, L, Greenblatt, H.M, Bageshwar, U.K, Savchenko, T, Gokhman, I, Sussman, J.L, Zamir, A, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2004-12-10 | | Release date: | 2005-05-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Three-dimensional structure of a halotolerant algal carbonic anhydrase predicts halotolerance of a mammalian homolog.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZGB
 
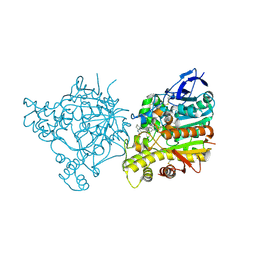 | | Crystal Structure of Torpedo Californica Acetylcholinesterase in Complex With an (R)-Tacrine(10)-Hupyridone Inhibitor. | | Descriptor: | (5R)-5-{[10-(1,2,3,4-TETRAHYDROACRIDIN-9-YLAMINO)DECYL]AMINO}-5,6,7,8-TETRAHYDROQUINOLIN-2(1H)-ONE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Haviv, H, Wong, D.M, Greenblatt, H.M, Carlier, P.R, Pang, Y.P, Silman, I, Sussman, J.L, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2005-04-21 | | Release date: | 2005-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Packing Mediates Enantioselective Ligand Recognition at the Peripheral Site of Acetylcholinesterase
J.Am.Chem.Soc., 127, 2005
|
|
1S0W
 
 | |
1U65
 
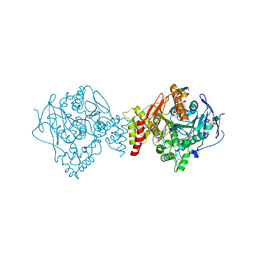 | | Ache W. CPT-11 | | Descriptor: | (4S)-4,11-DIETHYL-4-HYDROXY-3,14-DIOXO-3,4,12,14-TETRAHYDRO-1H-PYRANO[3',4':6,7]INDOLIZINO[1,2-B]QUINOLIN-9-YL 1,4'-BIPIPERIDINE-1'-CARBOXYLATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Harel, M, Hyatt, J.L, Brumshtein, B, Morton, C.L, Wadkins, R.W, Silman, I, Sussman, J.L, Potter, P.M, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2004-07-29 | | Release date: | 2005-07-19 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The crystal structure of the complex of the anticancer prodrug 7-ethyl-10-[4-(1-piperidino)-1-piperidino]-carbonyloxycamptothecin (CPT-11) with Torpedo californica acetylcholinesterase provides a molecular explanation for its cholinergic action
Mol.Pharmacol., 67, 2005
|
|
2V3F
 
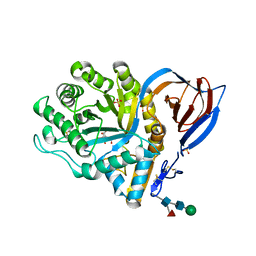 | | acid-beta-glucosidase produced in carrot | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLUCOSYLCERAMIDASE, SULFATE ION, ... | | Authors: | Shaaltiel, Y, Bartfeld, D, Hashmueli, S, Baum, G, Brill-Almon, E, Galili, G, Dym, O, Boldin-Adamsky, S.A, Silman, I, Sussman, J.L, Futerman, A.H, Aviezer, D, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2007-06-17 | | Release date: | 2008-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Production of Glucocerebrosidase with Terminal Mannose Glycans for Enzyme Replacement Therapy of Gaucher'S Disease Using a Plant Cell System.
Plant Biotechnol.J., 5, 2007
|
|
