1A0F
 
 | | CRYSTAL STRUCTURE OF GLUTATHIONE S-TRANSFERASE FROM ESCHERICHIA COLI COMPLEXED WITH GLUTATHIONESULFONIC ACID | | Descriptor: | GLUTATHIONE S-TRANSFERASE, GLUTATHIONE SULFONIC ACID | | Authors: | Nishida, M, Harada, S, Noguchi, S, Inoue, H, Takahashi, K, Satow, Y. | | Deposit date: | 1997-11-29 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structure of Escherichia coli glutathione S-transferase complexed with glutathione sulfonate: catalytic roles of Cys10 and His106.
J.Mol.Biol., 281, 1998
|
|
1GCQ
 
 | | CRYSTAL STRUCTURE OF VAV AND GRB2 SH3 DOMAINS | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2, VAV PROTO-ONCOGENE | | Authors: | Nishida, M, Nagata, K, Hachimori, Y, Ogura, K, Inagaki, F. | | Deposit date: | 2000-08-08 | | Release date: | 2001-08-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Novel recognition mode between Vav and Grb2 SH3 domains.
EMBO J., 20, 2001
|
|
1GCP
 
 | | CRYSTAL STRUCTURE OF VAV SH3 DOMAIN | | Descriptor: | VAV PROTO-ONCOGENE | | Authors: | Nishida, M, Nagata, K, Hachimori, Y, Ogura, K, Inagaki, F. | | Deposit date: | 2000-08-08 | | Release date: | 2001-08-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel recognition mode between Vav and Grb2 SH3 domains.
EMBO J., 20, 2001
|
|
2QKS
 
 | |
1N9P
 
 | |
7PCE
 
 | | BurG (apo): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ketol-acid reductoisomerase, PHOSPHATE ION | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCC
 
 | | BurG in complex with Mg2+ and NAD+ (holo): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | GLYCEROL, Ketol-acid reductoisomerase, MAGNESIUM ION, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCG
 
 | | BurG (holo) in complex with cyclopropane-1,1-dicarboxylate (7): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | GLYCEROL, Ketol-acid reductoisomerase, MAGNESIUM ION, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCM
 
 | | BurG (holo) in complex with (Z)-2,3-dihydroxy-6-methyl-hept-2-enoate (13): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | (Z)-6-methyl-2,3-bis(oxidanyl)hept-2-enoic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Ketol-acid reductoisomerase, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCO
 
 | | BurG E232Q mutant (holo) in complex with 2R,3R-2,3-dihydroxy-6-methyl-heptanoate (12): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | (2R,3R)-6-methyl-2,3-bis(oxidanyl)heptanoic acid, GLYCEROL, Ketol-acid reductoisomerase, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCI
 
 | | BurG (holo) in complex with hydroxypyruvate-enol (8): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | (Z)-2,3-bis(oxidanyl)prop-2-enoic acid, IMIDAZOLE, Ketol-acid reductoisomerase, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCT
 
 | | BurG E232Q mutant (holo) in complex with enol-oxalacetate (15): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | (~{Z})-2-oxidanylbut-2-enedioic acid, GLYCEROL, Ketol-acid reductoisomerase, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-04 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCL
 
 | | BurG (holo) in complex with 2-hydroxy-2-(hydroxy(isopropyl)amino)acetate (11): Biosynthesis of cyclopropanolrings in bacterial toxins | | Descriptor: | (2S)-2-oxidanyl-2-[oxidanyl(propan-2-yl)amino]ethanoic acid, Ketol-acid reductoisomerase, MAGNESIUM ION, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCN
 
 | | BurG (holo) in complex with gonyenediol (14), trigonic acid (6) and DMS: Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | (2R)-2-oxidanyl-2-(1-oxidanylcyclopropyl)ethanoic acid, (METHYLSULFANYL)METHANE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
8AFZ
 
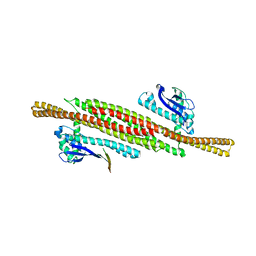 | | Architecture of the ESCPE-1 membrane coat | | Descriptor: | Cation-independent mannose-6-phosphate receptor, Sorting nexin-1, Sorting nexin-5 | | Authors: | Lopez-Robles, C, Scaramuzza, S, Astorga-Simon, E, Ishida, M, Williamsom, C.D, Banos-Mateos, S, Gil-Carton, D, Romero, M, Vidaurrazaga, A, Fernandez-Recio, J, Rojas, A.L, Bonifacino, J.S, Castano-Diez, D, Hierro, A. | | Deposit date: | 2022-07-18 | | Release date: | 2023-06-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Architecture of the ESCPE-1 membrane coat.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1V8Z
 
 | | X-ray crystal structure of the Tryptophan Synthase b2 Subunit from Hyperthermophile, Pyrococcus furiosus | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, Tryptophan synthase beta chain 1 | | Authors: | Hioki, Y, Ogasahara, K, Lee, S.J, Ma, J, Ishida, M, Yamagata, Y, Matsuura, Y, Ota, M, Kuramitsu, S, Yutani, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-15 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The crystal structure of the tryptophan synthase beta subunit from the hyperthermophile Pyrococcus furiosus. Investigation of stabilization factors
Eur.J.Biochem., 271, 2004
|
|
1WDW
 
 | | Structural basis of mutual activation of the tryptophan synthase a2b2 complex from a hyperthermophile, Pyrococcus furiosus | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Tryptophan synthase alpha chain, Tryptophan synthase beta chain 1 | | Authors: | Lee, S.J, Ogasahara, K, Ma, J, Nishio, K, Ishida, M, Yamagata, Y, Tsukihara, T, Yutani, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-19 | | Release date: | 2005-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Conformational Changes in the Tryptophan Synthase from a Hyperthermophile upon alpha(2)beta(2) Complex Formation: Crystal Structure of the Complex
Biochemistry, 44, 2005
|
|
3VND
 
 | | Crystal structure of tryptophan synthase alpha-subunit from the psychrophile Shewanella frigidimarina K14-2 | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, SULFATE ION, Tryptophan synthase alpha chain | | Authors: | Mitsuya, D, Tanaka, S, Matsumura, H, Takano, K, Urano, N, Ishida, M. | | Deposit date: | 2012-01-12 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Strategy for cold adaptation of the tryptophan synthase alpha subunit from the psychrophile Shewanella frigidimarina K14-2: crystal structure and physicochemical properties
J.Biochem., 155, 2014
|
|
8XZ2
 
 | | The structural model of a homodimeric D-Ala-D-Ala metallopeptidase, VanX, from vancomycin-resistant bacteria | | Descriptor: | D-alanyl-D-alanine dipeptidase | | Authors: | Konuma, T, Takai, T, Tsuchiya, C, Nishida, M, Hashiba, M, Yamada, Y, Shirai, H, Motoda, Y, Nagadoi, A, Chikaishi, E, Akagi, K, Akashi, S, Yamazaki, T, Akutsu, H, Oe, A, Ikegami, T. | | Deposit date: | 2024-01-20 | | Release date: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Analysis of the homodimeric structure of a D-Ala-D-Ala metallopeptidase, VanX, from vancomycin-resistant bacteria.
Protein Sci., 33, 2024
|
|
1K1Z
 
 | | Solution structure of N-terminal SH3 domain mutant(P33G) of murine Vav | | Descriptor: | vav | | Authors: | Ogura, K, Nagata, K, Horiuchi, M, Ebisui, E, Hasuda, T, Yuzawa, S, Nishida, M, Hatanaka, H, Inagaki, F. | | Deposit date: | 2001-09-26 | | Release date: | 2001-10-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of N-terminal SH3 domain of Vav and the recognition site for Grb2 C-terminal SH3 domain
J.BIOMOL.NMR, 22, 2002
|
|
2D5R
 
 | | Crystal Structure of a Tob-hCaf1 Complex | | Descriptor: | CCR4-NOT transcription complex subunit 7, Tob1 protein | | Authors: | Horiuchi, M, Suzuki, N.N, Muroya, N, Takahasi, K, Nishida, M, Yoshida, Y, Ikematsu, N, Nakamura, T, Kawamura-Tsuzuku, J, Yamamoto, T, Inagaki, F. | | Deposit date: | 2005-11-04 | | Release date: | 2006-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the antiproliferative activity of the Tob-hCaf1 complex.
J.Biol.Chem., 284, 2009
|
|
3TK5
 
 | | Factor Xa in complex with D102-4380 | | Descriptor: | 4-{3-[(4-chlorophenyl)amino]-3-oxopropyl}-3-({[5-(propan-2-yl)-4,5,6,7-tetrahydro[1,3]thiazolo[5,4-c]pyridin-2-yl]carbonyl}amino)benzoic acid, CALCIUM ION, Factor X heavy chain, ... | | Authors: | Suzuki, M, Mochizuki, A, Nagata, T, Takano, H, Kanno, H, Kishida, M, Ohta, T. | | Deposit date: | 2011-08-25 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Zwitter ionic potent durable orally active Factor Xa inhibitor.
To be Published
|
|
3TK6
 
 | | factor Xa in complex with D46-5241 | | Descriptor: | CALCIUM ION, Factor X heavy chain, Factor X light chain, ... | | Authors: | Suzuki, M, Mochizuki, A, Nagata, T, Takano, H, Kanno, H, Kishida, M, Ohta, T. | | Deposit date: | 2011-08-25 | | Release date: | 2012-08-29 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Zwitter ionic potent durable orally active Factor Xa inhibitor.
To be Published
|
|
