9AUE
 
 | | Crystal structure of the holo form of GenB2 in complex with PMP | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 6'-epimerase, C-6' aminotransferase, ... | | Authors: | Oliveira, G.S, Bury, P.S, Huang, F, Li, Y, Araujo, N.C, Zhou, J, Sun, Y, Leeper, F, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2024-02-29 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural and Functional Basis of GenB2 Isomerase Activity from Gentamicin Biosynthesis.
Acs Chem.Biol., 19, 2024
|
|
1LD9
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF AN H-2LD PEPTIDE COMPLEX EXPLAINS THE UNIQUE INTERACTION OF LD WITH BETA2M AND PEPTIDE | | Descriptor: | BETA-2 MICROGLOBULIN, MHC CLASS I H-2LD HEAVY CHAIN, NANO-PEPTIDE | | Authors: | Balendiran, G.K, Solheim, J.C, Young, A.C.M, Hansen, T.H, Nathenson, S.G, Sacchettini, J.C. | | Deposit date: | 1997-04-24 | | Release date: | 1998-05-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structure of an H-2Ld-peptide complex explains the unique interaction of Ld with beta-2 microglobulin and peptide.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
8ENT
 
 | | Interleukin-21 signaling complex with IL-21R and IL-2Rg | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Abhiraman, G.C, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-09-30 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | A structural blueprint for interleukin-21 signal modulation.
Cell Rep, 42, 2023
|
|
6N5F
 
 | | Crystal structure of an epoxide hydrolase from Trichoderma reesei in complex with inhibitor 3 | | Descriptor: | Epoxide hydrolase TrEH, N-(8-amino-8-oxooctyl)nonanamide | | Authors: | Oliveira, G.S, Adriani, P.P, Ribeiro, J.A, Morisseau, C, Hammock, B.D, Dias, M.V, Chambergo, F.S. | | Deposit date: | 2018-11-21 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The molecular structure of an epoxide hydrolase from Trichoderma reesei in complex with urea or amide-based inhibitors.
Int. J. Biol. Macromol., 129, 2019
|
|
6N5G
 
 | | Crystal structure of an epoxide hydrolase from Trichoderma reesei in complex with inhibitor 2 | | Descriptor: | 4-[(quinolin-3-yl)methyl]-N-[4-(trifluoromethoxy)phenyl]piperidine-1-carboxamide, Epoxide hydrolase TrEH | | Authors: | Oliveira, G.S, Adriani, P.P, Ribeiro, J.A, Morisseau, C, Hammock, B.D, Dias, M.V, Chambergo, F.S. | | Deposit date: | 2018-11-21 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The molecular structure of an epoxide hydrolase from Trichoderma reesei in complex with urea or amide-based inhibitors.
Int. J. Biol. Macromol., 129, 2019
|
|
6N5H
 
 | | Crystal structure of an epoxide hydrolase from Trichoderma reesei in complex with inhibitor 5 | | Descriptor: | 4-[(trans-4-{[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]dec-1-ylcarbamoyl]amino}cyclohexyl)oxy]benzoic acid, Epoxide hydrolase TrEH | | Authors: | Oliveira, G.S, Adriano, P.P, Ribeiro, J.A, Morisseau, C, Hammock, B.D, Dias, M.V, Chambergo, F.S. | | Deposit date: | 2018-11-21 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | The molecular structure of an epoxide hydrolase from Trichoderma reesei in complex with urea or amide-based inhibitors
Int. J. Biol. Macromol., 129, 2019
|
|
1FE3
 
 | |
1D6N
 
 | | TERNARY COMPLEX STRUCTURE OF HUMAN HGPRTASE, PRPP, MG2+, AND THE INHIBITOR HPP REVEALS THE INVOLVEMENT OF THE FLEXIBLE LOOP IN SUBSTRATE BINDING | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 3H-PYRAZOLO[4,3-D]PYRIMIDIN-7-OL, MAGNESIUM ION, ... | | Authors: | Balendiran, G.K. | | Deposit date: | 1999-10-14 | | Release date: | 1999-12-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ternary complex structure of human HGPRTase, PRPP, Mg2+, and the inhibitor HPP reveals the involvement of the flexible loop in substrate binding.
Protein Sci., 8, 1999
|
|
1FDQ
 
 | | CRYSTAL STRUCTURE OF HUMAN BRAIN FATTY ACID BINDING PROTEIN | | Descriptor: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, FATTY ACID-BINDING PROTEIN, BRAIN | | Authors: | Balendiran, G.K. | | Deposit date: | 2000-07-20 | | Release date: | 2001-07-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and thermodynamic analysis of human brain fatty acid-binding protein.
J.Biol.Chem., 275, 2000
|
|
6E49
 
 | | Pif1 peptide bound to PCNA trimer | | Descriptor: | ATP-dependent DNA helicase PIF1, Proliferating cell nuclear antigen | | Authors: | Buzovetsky, O, Kwon, Y, Pham, N.T, Kim, C, Ira, G, Sung, P, Xiong, Y. | | Deposit date: | 2018-07-17 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Role of the Pif1-PCNA Complex in Pol delta-Dependent Strand Displacement DNA Synthesis and Break-Induced Replication.
Cell Rep, 21, 2017
|
|
1H7J
 
 | | Solution structure of the 26 aa presequence of 5-ALAS | | Descriptor: | AMINOLEVULINIC ACID SYNTHASE 2, ERYTHROID | | Authors: | Goodfellow, B.J, Dias, J.S, Ferreira, G.C, Wray, V, Henklein, P, Macedo, A.L. | | Deposit date: | 2001-07-08 | | Release date: | 2001-10-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure and Heme Binding of the Presequence of Murine 5-Aminolevulinate Synthase
FEBS Lett., 505, 2001
|
|
1QF2
 
 | | THERMOLYSIN (E.C.3.4.24.27) COMPLEXED WITH (2-SULPHANYL-3-PHENYLPROPANOYL)-GLY-(5-PHENYLPROLINE). PARAMETERS FOR ZN-MONODENTATION OF MERCAPTOACYLDIPEPTIDES IN METALLOENDOPEPTIDASE | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, PROTEIN (THERMOLYSIN), ... | | Authors: | Gaucher, J.-F, Selkti, M, Tiraboschi, G, Prange, T, Roques, B.P, Tomas, A, Fournie-Zaluski, M.C. | | Deposit date: | 1999-04-06 | | Release date: | 1999-12-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structures of alpha-mercaptoacyldipeptides in the thermolysin active site: structural parameters for a Zn monodentation or bidentation in metalloendopeptidases.
Biochemistry, 38, 1999
|
|
2B5A
 
 | | C.BclI, Control Element of the BclI Restriction-Modification System | | Descriptor: | ACETIC ACID, C.BclI | | Authors: | Sawaya, M.R, Zhu, Z, Mersha, F, Chan, S.H, Dabur, R, Xu, S.Y, Balendiran, G.K. | | Deposit date: | 2005-09-28 | | Release date: | 2006-01-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | Crystal Structure of the Restriction-Modification System Control Element C.BclI and Mapping of Its Binding Site.
Structure, 13, 2005
|
|
2AC2
 
 | | Crystal structure of the Tyr13Phe mutant variant of Bacillus subtilis Ferrochelatase with Zn(2+) bound at the active site | | Descriptor: | Ferrochelatase, ZINC ION | | Authors: | Shipovskov, S, Karlberg, T, Fodje, M, Hansson, M.D, Ferreira, G.C, Hansson, M, Reimann, C.T, Al-Karadaghi, S. | | Deposit date: | 2005-07-18 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Metallation of the Transition-state Inhibitor N-methyl Mesoporphyrin by Ferrochelatase: Implications for the Catalytic Reaction Mechanism.
J.Mol.Biol., 352, 2005
|
|
2AC4
 
 | | Crystal structure of the His183Cys mutant variant of Bacillus subtilis Ferrochelatase | | Descriptor: | Ferrochelatase | | Authors: | Shipovskov, S, Karlberg, T, Fodje, M, Hansson, M.D, Ferreira, G.C, Hansson, M, Reimann, C.T, Al-Karadaghi, S. | | Deposit date: | 2005-07-18 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Metallation of the Transition-state Inhibitor N-methyl Mesoporphyrin by Ferrochelatase: Implications for the Catalytic Reaction Mechanism.
J.Mol.Biol., 352, 2005
|
|
9AU3
 
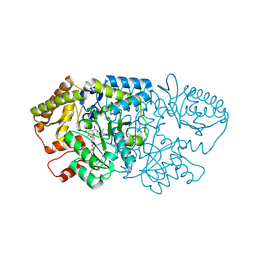 | | Crystal structure of GenB2 in complex with G418 | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 6'-epimerase, C-6' aminotransferase, ... | | Authors: | De Oliveira, G.S, Bury, P.S, Huang, F, Li, Y, Araujo, N.C, Zhou, J, Sun, Y, Leeper, F, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2024-02-28 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and Functional Basis of GenB2 Isomerase Activity from Gentamicin Biosynthesis.
Acs Chem.Biol., 19, 2024
|
|
9B0C
 
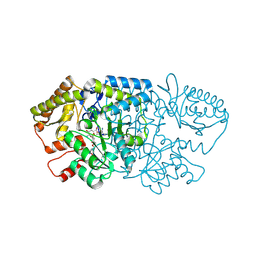 | | Crystal structure of GenB2 in complex with gentamicin X2. | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-diamino-3-{[3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranosyl]oxy}-2-hydroxycyclohexyl 2-amino-2-deoxy-alpha-D-glucopyranoside, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 6'-epimerase, ... | | Authors: | Bury, P.S, Araujo, N.C, Oliveira, G.S, Dias, M.V.B. | | Deposit date: | 2024-03-11 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural and Functional Basis of GenB2 Isomerase Activity from Gentamicin Biosynthesis.
Acs Chem.Biol., 19, 2024
|
|
3ST1
 
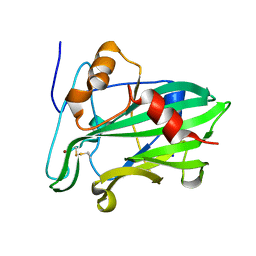 | | Crystal structure of Necrosis and Ethylene inducing Protein 2 from the causal agent of cocoa's Witches Broom disease | | Descriptor: | Necrosis-and ethylene-inducing protein, SODIUM ION, ZINC ION | | Authors: | Oliveira, J.F, Zaparoli, G, Barsottini, M.R.O, Ambrosio, A.L.B, Pereira, G.A.G, Dias, S.M.G. | | Deposit date: | 2011-07-08 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of Necrosis- and Ethylene-Inducing Protein 2 from the Causal Agent of Cacao's Witches' Broom Disease Reveals Key Elements for Its Activity.
Biochemistry, 50, 2011
|
|
3SUL
 
 | | Crystal structure of cerato-platanin 3 from M. perniciosa (MpCP3) | | Descriptor: | Cerato-platanin-like protein | | Authors: | Oliveira, J.F, Barsottini, M.R.O, Zaparoli, G, Machado, L.O, Dias, S.M.G, Pereira, G.A.G, Ambrosio, A.L.B. | | Deposit date: | 2011-07-11 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Functional diversification of cerato-platanins in Moniliophthora perniciosa as seen by differential expression and protein function specialization.
Mol. Plant Microbe Interact., 26, 2013
|
|
3SUJ
 
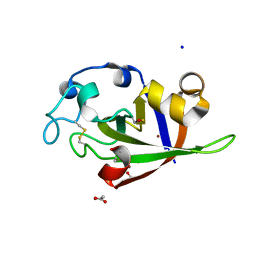 | | Crystal structure of cerato-platanin 1 from M. perniciosa (MpCP1) | | Descriptor: | ACETATE ION, CHLORIDE ION, Cerato-platanin 1, ... | | Authors: | Oliveira, J.F, Barsottini, M.R.O, Zaparoli, G, Machado, L.O, Dias, S.M.G, Pereira, G.A.G, Ambrosio, A.L.B. | | Deposit date: | 2011-07-11 | | Release date: | 2012-07-11 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Functional diversification of cerato-platanins in Moniliophthora perniciosa as seen by differential expression and protein function specialization.
Mol. Plant Microbe Interact., 26, 2013
|
|
3SUK
 
 | | Crystal structure of cerato-platanin 2 from M. perniciosa (MpCP2) | | Descriptor: | Cerato-platanin-like protein | | Authors: | Oliveira, J.F, Barsottini, M.R.O, Zaparoli, G, Machado, L.O, Dias, S.M.G, Pereira, G.A.G, Ambrosio, A.L.B. | | Deposit date: | 2011-07-11 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Functional diversification of cerato-platanins in Moniliophthora perniciosa as seen by differential expression and protein function specialization.
Mol. Plant Microbe Interact., 26, 2013
|
|
3SUM
 
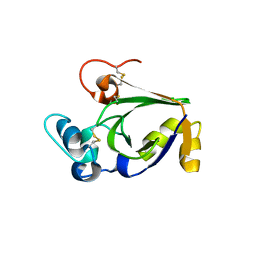 | | Crystal structure of cerato-platanin 5 from M. perniciosa (MpCP5) | | Descriptor: | Cerato-platanin-like protein | | Authors: | Oliveira, J.F, Barsottini, M.R.O, Zaparoli, G, Machado, L.O, Dias, S.M.G, Pereira, G.A.G, Ambrosio, A.L.B. | | Deposit date: | 2011-07-11 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Functional diversification of cerato-platanins in Moniliophthora perniciosa as seen by differential expression and protein function specialization.
Mol. Plant Microbe Interact., 26, 2013
|
|
4H37
 
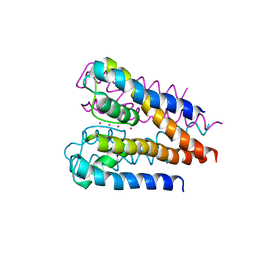 | | Crystal structure of a voltage-gated K+ channel pore domain in a closed state in lipid membranes | | Descriptor: | Lmo2059 protein, POTASSIUM ION | | Authors: | Santos, J.S, Asmar-Rovira, G.A, Han, G.W, Liu, W, Syeda, R, Cherezov, V, Baker, K.A, Stevens, R.C, Montal, M. | | Deposit date: | 2012-09-13 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal Structure of a Voltage-gated K+ Channel Pore Module in a Closed State in Lipid Membranes.
J.Biol.Chem., 287, 2012
|
|
4H33
 
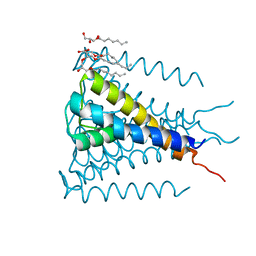 | | Crystal structure of a voltage-gated K+ channel pore module in a closed state in lipid membranes, tetragonal crystal form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Lmo2059 protein, POTASSIUM ION | | Authors: | Santos, J.S, Asmar-Rovira, G.A, Han, G.W, Liu, W, Syeda, R, Cherezov, V, Baker, K.A, Stevens, R.C, Montal, M. | | Deposit date: | 2012-09-13 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of a Voltage-gated K+ Channel Pore Module in a Closed State in Lipid Membranes.
J.Biol.Chem., 287, 2012
|
|
4Z36
 
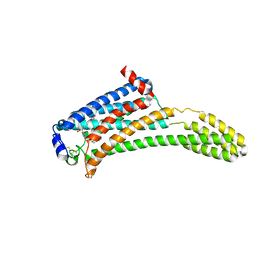 | | Crystal Structure of Human Lysophosphatidic Acid Receptor 1 in complex with ONO-3080573 | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 1-(4-{[(2S,3R)-2-(2,3-dihydro-1H-inden-2-yloxy)-3-(3,5-dimethoxy-4-methylphenyl)-3-hydroxypropyl]oxy}phenyl)cyclopropanecarboxylic acid, Lysophosphatidic acid receptor 1,Soluble cytochrome b562 | | Authors: | Chrencik, J.E, Roth, C.B, Terakado, M, Kurata, H, Omi, R, Kihara, Y, Warshaviak, D, Nakade, S, Asmar-Rovira, G, Mileni, M, Mizuno, H, Griffith, M.T, Rodgers, C, Han, G.W, Velasquez, J, Chun, J, Stevens, R.C, Hanson, M.A, GPCR Network (GPCR) | | Deposit date: | 2015-03-30 | | Release date: | 2015-06-03 | | Last modified: | 2015-07-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1.
Cell, 161, 2015
|
|
