4D10
 
 | | Crystal structure of the COP9 signalosome | | 分子名称: | COP9 SIGNALOSOME COMPLEX SUBUNIT 1, COP9 SIGNALOSOME COMPLEX SUBUNIT 2, COP9 SIGNALOSOME COMPLEX SUBUNIT 3, ... | | 著者 | Bunker, R.D, Lingaraju, G.M, Thoma, N.H. | | 登録日 | 2014-04-30 | | 公開日 | 2014-07-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Crystal Structure of the Human Cop9 Signalosome
Nature, 512, 2014
|
|
6AZA
 
 | |
5DG6
 
 | | 2.35A resolution structure of Norovirus 3CL protease in complex an oxadiazole-based, cell permeable macrocyclic (21-mer) inhibitor | | 分子名称: | 3C-LIKE PROTEASE, CHLORIDE ION, tert-butyl [(4S,7S,10S)-7-(cyclohexylmethyl)-10-(hydroxymethyl)-5,8,13-trioxo-23-oxa-6,9,14,21,22-pentaazabicyclo[18.2.1]tricosa-1(22),20-dien-4-yl]carbamate | | 著者 | Lovell, S, Battaile, K.P, Mehzabeen, N, Damalanka, V.C, Kim, Y, Alliston, K.R, Weerawarna, P.M, Kankanamalage, A.C.G, Lushington, G.H, Chang, K.-O, Groutas, W.C. | | 登録日 | 2015-08-27 | | 公開日 | 2016-02-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Oxadiazole-Based Cell Permeable Macrocyclic Transition State Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 59, 2016
|
|
1D8M
 
 | | CRYSTAL STRUCTURE OF MMP3 COMPLEXED WITH A HETEROCYCLE-BASED INHIBITOR | | 分子名称: | 1-BENZYL-3-(4-METHOXY-BENZENESULFONYL)-6-OXO-HEXAHYDRO-PYRIMIDINE-4-CARBOXYLIC ACID HYDROXYAMIDE, CALCIUM ION, STROMELYSIN-1 PRECURSOR, ... | | 著者 | Pikul, S, Dunham, K.M, Almstead, N.G, De, B, Natchus, M.G, Taiwo, Y.O, Williams, L.E, Hynd, B.A, Hsieh, L.C, Janusz, M.J, Gu, F, Mieling, G.E. | | 登録日 | 1999-10-25 | | 公開日 | 2000-10-25 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Heterocycle-based MMP inhibitors with P2' substituents.
Bioorg.Med.Chem.Lett., 11, 2001
|
|
3L28
 
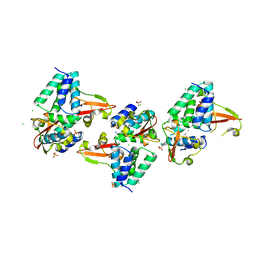 | | Crystal structure of Zaire Ebola VP35 interferon inhibitory domain K339A mutant | | 分子名称: | CHLORIDE ION, Polymerase cofactor VP35, SODIUM ION, ... | | 著者 | Leung, D.W, Prins, K.C, Borek, D.M, Farahbakhsh, M, Tufariello, J.M, Ramanan, P, Nix, J.C, Helgeson, L.A, Otwinowski, Z, Honzatko, R.B, Basler, C.F, Amarasinghe, G.K. | | 登録日 | 2009-12-14 | | 公開日 | 2010-01-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5DGJ
 
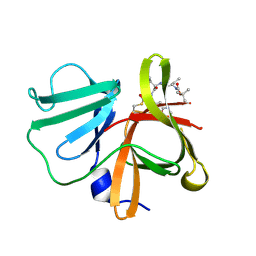 | | 1.0A resolution structure of Norovirus 3CL protease in complex an oxadiazole-based, cell permeable macrocyclic (20-mer) inhibitor | | 分子名称: | 3C-LIKE PROTEASE, tert-butyl [(4S,7S,10S)-7-(cyclohexylmethyl)-10-(hydroxymethyl)-5,8,13-trioxo-22-oxa-6,9,14,20,21-pentaazabicyclo[17.2.1]docosa-1(21),19-dien-4-yl]carbamate | | 著者 | Lovell, S, Battaile, K.P, Mehzabeen, N, Damalanka, V.C, Kim, Y, Alliston, K.R, Weerawarna, P.M, Kankanamalage, A.C.G, Lushington, G.H, Chang, K.-O, Groutas, W.C. | | 登録日 | 2015-08-27 | | 公開日 | 2016-02-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Oxadiazole-Based Cell Permeable Macrocyclic Transition State Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 59, 2016
|
|
3L2A
 
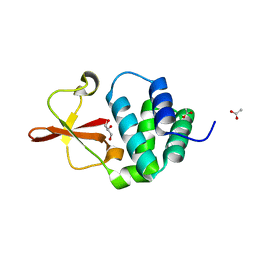 | | Crystal structure of Reston Ebola VP35 interferon inhibitory domain | | 分子名称: | ACETIC ACID, GLYCEROL, Polymerase cofactor VP35 | | 著者 | Leung, D.W, Farahbakhsh, M, Borek, D.M, Prins, K.C, Basler, C.F, Amarasinghe, G.K. | | 登録日 | 2009-12-14 | | 公開日 | 2010-05-12 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Structural and Functional Characterization of Reston Ebola Virus VP35 Interferon Inhibitory Domain.
J.Mol.Biol., 399, 2010
|
|
7LDK
 
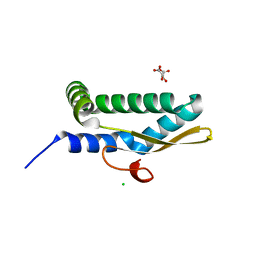 | | Structure of human respiratory syncytial virus nonstructural protein 2 (NS2) | | 分子名称: | CHLORIDE ION, D(-)-TARTARIC ACID, Non-structural protein 2 | | 著者 | Chatterjee, S, Borek, D, Otwinowski, Z, Amarasinghe, G.K, Leung, D.W. | | 登録日 | 2021-01-13 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.82 Å) | | 主引用文献 | Structural basis for IFN antagonism by human respiratory syncytial virus nonstructural protein 2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4CDU
 
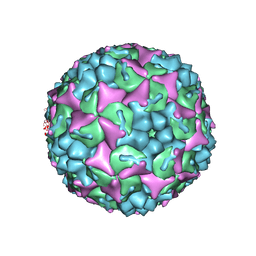 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP3 | | 分子名称: | 1-[(3S)-5-[4-[(E)-ETHOXYIMINOMETHYL]PHENOXY]-3-METHYL-PENTYL]-3-PYRIDIN-4-YL-IMIDAZOLIDIN-2-ONE, CHLORIDE ION, SODIUM ION, ... | | 著者 | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | 登録日 | 2013-11-06 | | 公開日 | 2014-02-12 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
5BU5
 
 | |
4CDQ
 
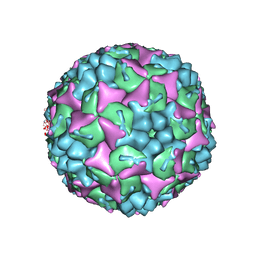 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP2 | | 分子名称: | 4-((5-(2-oxo-3-(pyridin-4-yl)imidazolidin-1-yl)pentyl)oxy)benzaldehyde O-ethyl oxime, SODIUM ION, VP1, ... | | 著者 | DeColibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | 登録日 | 2013-11-05 | | 公開日 | 2014-02-12 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
4EI2
 
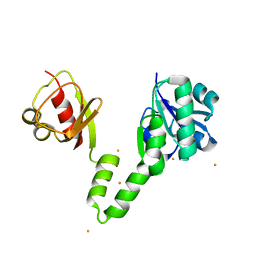 | |
5ABJ
 
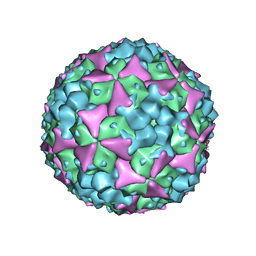 | | Structure of Coxsackievirus A16 in complex with GPP3 | | 分子名称: | 1-[(3S)-5-[4-[(E)-ETHOXYIMINOMETHYL]PHENOXY]-3-METHYL-PENTYL]-3-PYRIDIN-4-YL-IMIDAZOLIDIN-2-ONE, CHLORIDE ION, SODIUM ION, ... | | 著者 | De Colibus, L, Wang, X, Tijsma, A, Neyts, J, Spyrou, J.A.B, Ren, J, Grimes, J.M, Puerstinger, G, Leyssen, P, Fry, E.E, Rao, Z, Stuart, D.I. | | 登録日 | 2015-08-06 | | 公開日 | 2015-09-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structure Elucidation of Coxsackievirus A16 in Complex with Gpp3 Informs a Systematic Review of Highly Potent Capsid Binders to Enteroviruses.
Plos Pathog., 11, 2015
|
|
1TUH
 
 | | Structure of Bal32a from a Soil-Derived Mobile Gene Cassette | | 分子名称: | ACETATE ION, hypothetical protein EGC068 | | 著者 | Robinson, A, Wu, P.S.-C, Harrop, S.J, Schaeffer, P.M, Dixon, N.E, Gillings, M.R, Holmes, A.J, Nevalainen, K.M.H, Otting, G, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C. | | 登録日 | 2004-06-25 | | 公開日 | 2004-07-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Integron-associated Mobile Gene Cassettes Code for Folded Proteins: The Structure of Bal32a, a New Member of the Adaptable alpha+beta Barrel Family
J.Mol.Biol., 346, 2005
|
|
6AY6
 
 | | Naegleria fowleri CYP51-voriconazole complex | | 分子名称: | CYP51, sterol 14alpha-demethylase, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Debnath, A, Calvet, C.M, Jennings, G, Zhou, W, Aksenov, A, Luth, M, Abagyan, R, Nes, W.D, McKerrow, J.H, Podust, L.M. | | 登録日 | 2017-09-07 | | 公開日 | 2017-11-22 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | CYP51 is an essential drug target for the treatment of primary amoebic meningoencephalitis (PAM).
PLoS Negl Trop Dis, 11, 2017
|
|
4H8S
 
 | |
3L29
 
 | | Crystal Structure of Zaire Ebola VP35 interferon inhibitory domain K319A/R322A mutant | | 分子名称: | CHLORIDE ION, Polymerase cofactor VP35 | | 著者 | Leung, D.W, Ramanan, P, Borek, D.M, Amarasinghe, G.K. | | 登録日 | 2009-12-14 | | 公開日 | 2010-02-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Mutations abrogating VP35 interaction with double-stranded RNA render ebola virus avirulent in guinea pigs.
J.Virol., 84, 2010
|
|
3L27
 
 | | Crystal structure of Zaire Ebola VP35 interferon inhibitory domain R312A mutant | | 分子名称: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | 著者 | Leung, D.W, Prins, K.C, Borek, D.M, Farahbakhsh, M, Tufariello, J.M, Ramanan, P, Nix, J.C, Helgeson, L.A, Otwinowski, Z, Honzatko, R.B, Basler, C.F, Amarasinghe, G.K. | | 登録日 | 2009-12-14 | | 公開日 | 2010-01-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3L26
 
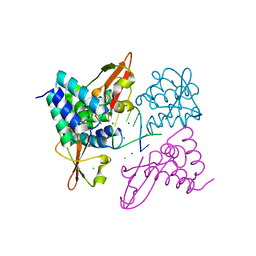 | | Crystal structure of Zaire Ebola VP35 interferon inhibitory domain bound to 8 bp dsRNA | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, Polymerase cofactor VP35, ... | | 著者 | Leung, D.W, Prins, K.C, Borek, D.M, Farahbakhsh, M, Tufariello, J.M, Ramanan, P, Nix, J.C, Helgeson, L.A, Otwinowski, Z, Honzatko, R.B, Basler, C.F, Amarasinghe, G.K. | | 登録日 | 2009-12-14 | | 公開日 | 2010-01-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5BU8
 
 | |
3LQF
 
 | | Crystal structure of the short-chain dehydrogenase Galactitol-Dehydrogenase (GatDH) of Rhodobacter sphaeroides in complex with NAD and erythritol | | 分子名称: | Galactitol dehydrogenase, MAGNESIUM ION, MESO-ERYTHRITOL, ... | | 著者 | Carius, Y, Christian, H, Faust, A, Kornberger, P, Zander, U, Klink, B.U, Kohring, G.W, Giffhorn, F, Scheidig, A.J. | | 登録日 | 2010-02-09 | | 公開日 | 2010-04-21 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural insight into substrate differentiation of the sugar-metabolizing enzyme galactitol dehydrogenase from Rhodobacter sphaeroides D.
J.Biol.Chem., 285, 2010
|
|
6MZT
 
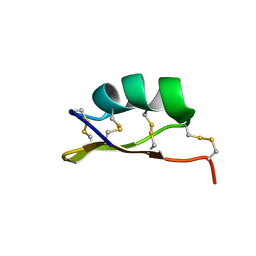 | | Solution structure of alpha-KTx-6.21 (UroTx) from Urodacus yaschenkoi | | 分子名称: | Potassium channel toxin alpha-KTx 6.21 | | 著者 | Chin, Y.K.-Y, Luna-Ramirez, K, Anangi, R, King, G.F. | | 登録日 | 2018-11-05 | | 公開日 | 2020-03-11 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis of the potency and selectivity of Urotoxin, a potent Kv1 blocker from scorpion venom.
Biochem. Pharmacol., 174, 2020
|
|
7RFD
 
 | |
5E0G
 
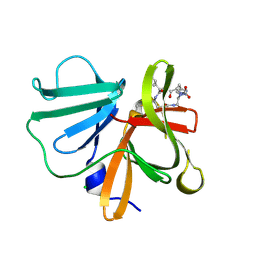 | | 1.20 A resolution structure of Norovirus 3CL protease in complex with a triazole-based macrocyclic (17-mer) inhibitor | | 分子名称: | (phenylmethyl) ~{N}-[(8~{S},11~{S},14~{S})-8-(hydroxymethyl)-11-(2-methylpropyl)-5,10,13-tris(oxidanylidene)-1,4,9,12,17,18-hexazabicyclo[14.2.1]nonadeca-16(19),17-dien-14-yl]carbamate, CHLORIDE ION, Norovirus 3C-like protease | | 著者 | Lovell, S, Battaile, K.P, Mehzabeen, N, Weerawarna, P.M, Kim, Y, Kankanamalage, A.C.G, Damalanka, V.C, Lushington, G.H, Alliston, K.R, Chang, K.-O, Groutas, W.C. | | 登録日 | 2015-09-28 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structure-based design and synthesis of triazole-based macrocyclic inhibitors of norovirus protease: Structural, biochemical, spectroscopic, and antiviral studies.
Eur.J.Med.Chem., 119, 2016
|
|
4K6B
 
 | |
