3OAA
 
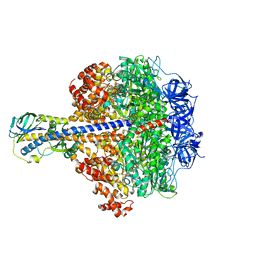 | |
4QLP
 
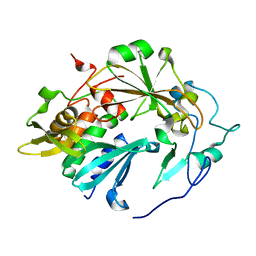 | | Atomic structure of tuberculosis necrotizing toxin (TNT) complexed with its immunity factor IFT | | Descriptor: | Alanine and proline rich protein, tuberculosis necrotizing toxin (TNT), immunity factor IFT | | Authors: | Cingolani, G, Lokareddy, R.K, Sun, J, Siroy, A, Speer, A, Doornbos, K.S, Niederweis, M. | | Deposit date: | 2014-06-12 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The tuberculosis necrotizing toxin kills macrophages by hydrolyzing NAD.
Nat.Struct.Mol.Biol., 22, 2015
|
|
6C6K
 
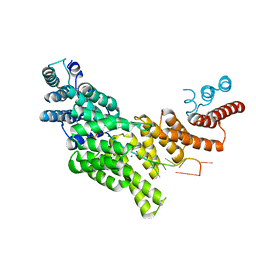 | | Structural basis for preferential recognition of cap 0 RNA by a human IFIT1-IFIT3 protein complex | | Descriptor: | Interferon-induced protein with tetratricopeptide repeats 1, Interferon-induced protein with tetratricopeptide repeats 3, MAGNESIUM ION, ... | | Authors: | Amarasinghe, G.K, Leung, D.W, Johnson, B, Xu, W. | | Deposit date: | 2018-01-18 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Human IFIT3 Modulates IFIT1 RNA Binding Specificity and Protein Stability.
Immunity, 48, 2018
|
|
7JOQ
 
 | | Structure of NV1 small terminase | | Descriptor: | Small Terminase subunit | | Authors: | Cingolani, G, Lokareddy, R. | | Deposit date: | 2020-08-07 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Biophysical analysis of Pseudomonas-phage PaP3 small terminase suggests a mechanism for sequence-specific DNA-binding by lateral interdigitation.
Nucleic Acids Res., 48, 2020
|
|
3QI5
 
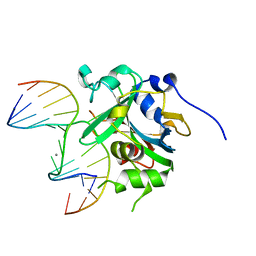 | | Crystal structure of human alkyladenine DNA glycosylase in complex with 3,N4-ethenocystosine containing duplex DNA | | Descriptor: | DNA (5'-D(*GP*AP*CP*AP*TP*GP*(EDC)P*TP*TP*GP*CP*CP*T)-3'), DNA (5'-D(*GP*GP*CP*AP*AP*GP*CP*AP*TP*GP*TP*CP*A)-3'), DNA-3-methyladenine glycosylase, ... | | Authors: | Lingaraju, G.M, Davis, C.A, Setser, J.W, Samson, L.D, Drennan, C.L. | | Deposit date: | 2011-01-26 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Inhibition of Human Alkyladenine DNA Glycosylase (AAG) by 3,N4-Ethenocytosine-containing DNA.
J.Biol.Chem., 286, 2011
|
|
4I1K
 
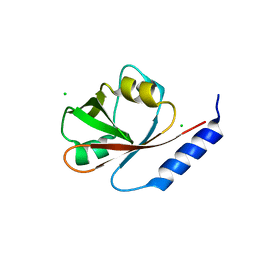 | | Crystal Structure of VRN1 (Residues 208-341) | | Descriptor: | B3 domain-containing transcription factor VRN1, CHLORIDE ION | | Authors: | King, G, Chanson, A.H, McCallum, E.J, Ohme-Takagi, M, Byriel, K, Hill, J.M, Martin, J.L, Mylne, J.S. | | Deposit date: | 2012-11-21 | | Release date: | 2012-12-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Arabidopsis B3 Domain Protein VERNALIZATION1 (VRN1) Is Involved in Processes Essential for Development, with Structural and Mutational Studies Revealing Its DNA-binding Surface.
J.Biol.Chem., 288, 2013
|
|
5U6X
 
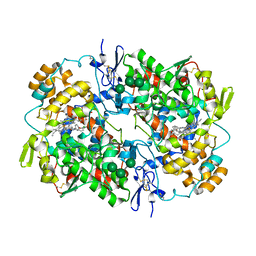 | | COX-1:P6 COMPLEX STRUCTURE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(5-chlorofuran-2-yl)-5-methyl-4-phenyl-1,2-oxazole, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Cingolani, G, Panella, A, Perrone, M.G, Vitale, P, Smith, W.L, Scilimati, A. | | Deposit date: | 2016-12-09 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structural basis for selective inhibition of Cyclooxygenase-1 (COX-1) by diarylisoxazoles mofezolac and 3-(5-chlorofuran-2-yl)-5-methyl-4-phenylisoxazole (P6).
Eur J Med Chem, 138, 2017
|
|
5VCL
 
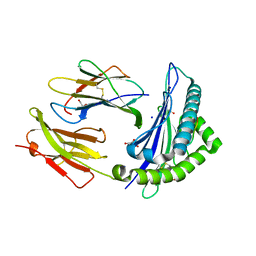 | | Structure of the Qdm peptide bound to Qa-1a | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H2-T23 protein, ... | | Authors: | Ying, G, Zajonc, D.M. | | Deposit date: | 2017-03-31 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Qa-1a with bound Qa-1 determinant modifier peptide.
PLoS ONE, 12, 2017
|
|
5W16
 
 | |
1G2X
 
 | | Sequence induced trimerization of krait PLA2: crystal structure of the trimeric form of krait PLA2 | | Descriptor: | PHOSPHOLIPASE A2 | | Authors: | Singh, G, Gourinath, S, Sharma, S, Bhanumathi, S, Paramsivam, M, Singh, T.P. | | Deposit date: | 2000-10-22 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sequence-induced trimerization of phospholipase A2: structure of a trimeric isoform of PLA2 from common krait (Bungarus caeruleus) at 2.5 A resolution.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1DPY
 
 | | THREE-DIMENSIONAL STRUCTURE OF A NOVEL PHOSPHOLIPASE A2 FROM INDIAN COMMON KRAIT AT 2.45 A RESOLUTION | | Descriptor: | PHOSPHOLIPASE A2, SODIUM ION | | Authors: | Singh, G, Gourinath, S, Sharma, S, Paramasivam, M, Srinivasan, A, Singh, T.P. | | Deposit date: | 1999-12-28 | | Release date: | 2000-06-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Sequence and crystal structure determination of a basic phospholipase A2 from common krait (Bungarus caeruleus) at 2.4 A resolution: identification and characterization of its pharmacological sites.
J.Mol.Biol., 307, 2001
|
|
6E7E
 
 | |
6E6A
 
 | | Triclinic crystal form of IncA G144A point mutant | | Descriptor: | Inclusion membrane protein A, SODIUM ION | | Authors: | Cingolani, G, Paumet, F. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the homotypic fusion of chlamydial inclusions by the SNARE-like protein IncA.
Nat Commun, 10, 2019
|
|
3FKE
 
 | | Structure of the Ebola VP35 Interferon Inhibitory Domain | | Descriptor: | Polymerase cofactor VP35 | | Authors: | Amarasinghe, G.K, Leung, D.W, Ginder, N.D, Honzatko, R.B, Nix, J, Basler, C.F, Fulton, D.B. | | Deposit date: | 2008-12-16 | | Release date: | 2009-01-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the Ebola VP35 interferon inhibitory domain.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6DLI
 
 | |
7UJ5
 
 | |
1M5N
 
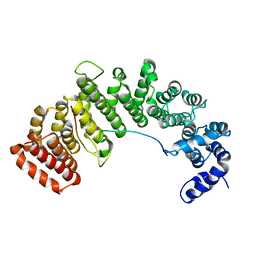 | | Crystal structure of HEAT repeats (1-11) of importin b bound to the non-classical NLS(67-94) of PTHrP | | Descriptor: | Importin beta-1 subunit, Parathyroid hormone-related protein | | Authors: | Cingolani, G, Bednenko, J, Gillespie, M.T, Gerace, L. | | Deposit date: | 2002-07-09 | | Release date: | 2003-01-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular basis for the recognition
of a nonclassical nuclear localization
signal by importin beta
Mol.Cell, 10, 2002
|
|
1PO8
 
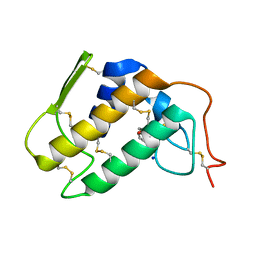 | | Crystal structure of a complex formed between krait venom phospholipase A2 and heptanoic acid at 2.7 A resolution. | | Descriptor: | HEPTANOIC ACID, Phospholipase A2, SODIUM ION | | Authors: | Singh, G, Jayasankar, J, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2003-06-14 | | Release date: | 2004-05-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of a complex formed between krait venom phospholipase A2 and heptanoic acid at 2.7 A resolution.
To be Published
|
|
1FE5
 
 | | SEQUENCE AND CRYSTAL STRUCTURE OF A BASIC PHOSPHOLIPASE A2 FROM COMMON KRAIT (BUNGARUS CAERULEUS) AT 2.4 RESOLUTION: IDENTIFICATION AND CHARACTERIZATION OF ITS PHARMACOLOGICAL SITES. | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Singh, G, Gourinath, S, Sharma, S, Paramasivam, M, Srinivasan, A, Singh, T.P. | | Deposit date: | 2000-07-21 | | Release date: | 2001-01-24 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Sequence and crystal structure determination of a basic phospholipase A2 from common krait (Bungarus caeruleus) at 2.4 A resolution: identification and characterization of its pharmacological sites.
J.Mol.Biol., 307, 2001
|
|
1EV0
 
 | |
1ESY
 
 | |
2M6J
 
 | |
2M35
 
 | | NMR study of k-Ssm1a | | Descriptor: | k-Ssm1a | | Authors: | King, G.F, Undheim, E.A, Mobli, M, Yang, S, Rong, M, Lai, R. | | Deposit date: | 2013-01-09 | | Release date: | 2014-01-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR study of k-Ssm1a
To be Published
|
|
2M36
 
 | |
1QGR
 
 | | STRUCTURE OF IMPORTIN BETA BOUND TO THE IBB DOMAIN OF IMPORTIN ALPHA (II CRYSTAL FORM, GROWN AT LOW PH) | | Descriptor: | PROTEIN (IMPORTIN ALPHA-2 SUBUNIT), PROTEIN (IMPORTIN BETA SUBUNIT) | | Authors: | Cingolani, G, Petosa, C, Weis, K, Muller, C.W. | | Deposit date: | 1999-05-04 | | Release date: | 1999-05-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of importin-beta bound to the IBB domain of importin-alpha.
Nature, 399, 1999
|
|
