5WBE
 
 | | COX-1:MOFEZOLAC COMPLEX STRUCTURE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mofezolac, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Cingolani, G, Panella, A, Perrone, M.G, Vitale, P, Smith, W.L, Scilimati, A. | | Deposit date: | 2017-06-28 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for selective inhibition of Cyclooxygenase-1 (COX-1) by diarylisoxazoles mofezolac and 3-(5-chlorofuran-2-yl)-5-methyl-4-phenylisoxazole (P6).
Eur J Med Chem, 138, 2017
|
|
6O4Z
 
 | | Structure of HLA-A2:01 with peptide MM92 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, MHC class I antigen, ... | | Authors: | Ying, G, Bitra, A, Zajonc, D.M. | | Deposit date: | 2019-03-01 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Anin silico-in vitroPipeline Identifying an HLA-A*02:01+KRAS G12V+Spliced Epitope Candidate for a Broad Tumor-Immune Response in Cancer Patients.
Front Immunol, 10, 2019
|
|
6O51
 
 | | Structure of HLA-A2:01 with peptide MM90 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, MHC class I antigen, ... | | Authors: | Ying, G, Bitra, A, Zajonc, D.M. | | Deposit date: | 2019-03-01 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Anin silico-in vitroPipeline Identifying an HLA-A*02:01+KRAS G12V+Spliced Epitope Candidate for a Broad Tumor-Immune Response in Cancer Patients.
Front Immunol, 10, 2019
|
|
4GUX
 
 | | Crystal structure of trypsin:MCoTi-II complex | | Descriptor: | ACETATE ION, CALCIUM ION, Cationic trypsin, ... | | Authors: | King, G.J, Daly, N.L, Thorstholm, L, Greenwood, K.P, Rosengren, K.J, Heras, B, Craik, D.J, Martin, J.L. | | Deposit date: | 2012-08-30 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structural insights into the role of the cyclic backbone in a squash trypsin inhibitor
J.Biol.Chem., 288, 2013
|
|
6O4Y
 
 | | Structure of HLA-A2:01 with peptide MM91 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, MHC class I antigen, ... | | Authors: | Ying, G, Bitra, A, Zajonc, D.M. | | Deposit date: | 2019-03-01 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Anin silico-in vitroPipeline Identifying an HLA-A*02:01+KRAS G12V+Spliced Epitope Candidate for a Broad Tumor-Immune Response in Cancer Patients.
Front Immunol, 10, 2019
|
|
6O53
 
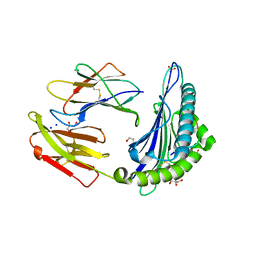 | | Structure of HLA-A2:01 with peptide MM96 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, MHC class I antigen, ... | | Authors: | Ying, G, Bitra, A, Zajonc, D.M. | | Deposit date: | 2019-03-01 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Anin silico-in vitroPipeline Identifying an HLA-A*02:01+KRAS G12V+Spliced Epitope Candidate for a Broad Tumor-Immune Response in Cancer Patients.
Front Immunol, 10, 2019
|
|
6OOR
 
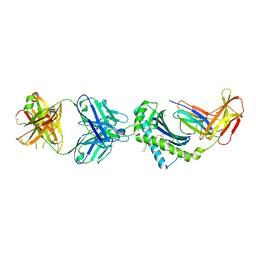 | | Structure of 1B1 bound to mouse CD1d | | Descriptor: | Antibody 1B1 Heavy chain, Antibody 1B1 Light chain, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Ying, G, Zajonc, D.M. | | Deposit date: | 2019-04-23 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis of NKT cell inhibition using the T-cell receptor-blocking anti-CD1d antibody 1B1.
J.Biol.Chem., 294, 2019
|
|
7M5C
 
 | |
6VI1
 
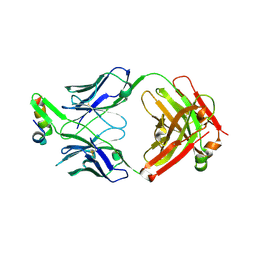 | | Structure of Fab4 bound to P22 TerL(1-33) | | Descriptor: | Synthetic Fab4 heavy chain, Synthetic Fab4 light chain, Terminase, ... | | Authors: | Cingolani, G, Lokareddy, R, Ko, Y. | | Deposit date: | 2020-01-11 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Recognition of an alpha-helical hairpin in P22 large terminase by a synthetic antibody fragment.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
3V00
 
 | |
7M5B
 
 | |
1TC8
 
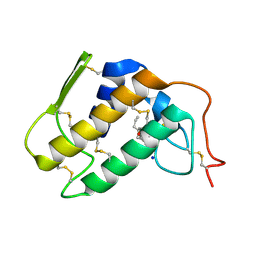 | | Crystal structure of Krait-venom phospholipase A2 in a complex with a natural fatty acid tridecanoic acid | | Descriptor: | N-TRIDECANOIC ACID, SODIUM ION, phospholipase A2 isoform 1 | | Authors: | Singh, G, Jasti, J, Saravanan, K, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2004-05-21 | | Release date: | 2004-06-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the complex formed between a group I phospholipase A2 and a naturally occurring fatty acid at 2.7 A resolution
PROTEIN SCI., 14, 2005
|
|
1U4J
 
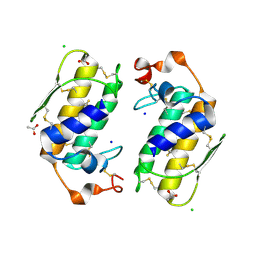 | | Crystal structure of a carbohydrate induced dimer of group I phospholipase A2 from Bungarus caeruleus at 2.1 A resolution | | Descriptor: | ACETIC ACID, CHLORIDE ION, SODIUM ION, ... | | Authors: | Singh, G, Gourinath, S, Sharma, S, Bhanumathi, S, Betzel, C, Srinivasan, A, Singh, T.P. | | Deposit date: | 2004-07-26 | | Release date: | 2004-08-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal structure of a carbohydrate induced homodimer of phospholipase A(2) from Bungarus caeruleus at 2.1A resolution
J.Struct.Biol., 149, 2005
|
|
7M5A
 
 | |
6XMI
 
 | | Structure of Fab4 bound to P22 TerL(1-33) | | Descriptor: | Fab Heavy chain, Fab Light chain, Terminase, ... | | Authors: | Cingolani, G, Lokareddy, R, Ko, Y. | | Deposit date: | 2020-06-30 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Recognition of an alpha-helical hairpin in P22 large terminase by a synthetic antibody fragment.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6VI2
 
 | | Structure of the unaligned Fab4 | | Descriptor: | FAB4 heavy chain, FAB4 light chain | | Authors: | Cingolani, G, Lokareddy, R, Ko, Y. | | Deposit date: | 2020-01-11 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Recognition of an alpha-helical hairpin in P22 large terminase by a synthetic antibody fragment.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6W7T
 
 | | Structure of PaP3 small terminase | | Descriptor: | small terminase subunit | | Authors: | Cingolani, G, Lokareddy, R. | | Deposit date: | 2020-03-19 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Biophysical analysis of Pseudomonas-phage PaP3 small terminase suggests a mechanism for sequence-specific DNA-binding by lateral interdigitation.
Nucleic Acids Res., 48, 2020
|
|
8DKR
 
 | |
1XQP
 
 | | Crystal structure of 8-oxoguanosine complexed Pa-AGOG, 8-oxoguanine DNA glycosylase from Pyrobaculum aerophilum | | Descriptor: | 2'-DEOXY-8-OXOGUANOSINE, 8-oxoguanine DNA glycosylase | | Authors: | Lingaraju, G.M, Sartori, A.A, Kostrewa, D, Prota, A.E, Jiricny, J, Winkler, F.K. | | Deposit date: | 2004-10-13 | | Release date: | 2004-11-16 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A DNA glycosylase from Pyrobaculum aerophilum with an 8-oxoguanine binding mode and a noncanonical helix-hairpin-helix structure
Structure, 13, 2005
|
|
8EB3
 
 | |
8VK3
 
 | |
8VJK
 
 | |
8VJJ
 
 | | Structure of mouse RyR1 (EGTA-only dataset) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Peptidyl-prolyl cis-trans isomerase FKBP1A, Ryanodine receptor 1, ... | | Authors: | Weninger, G, Marks, A.R. | | Deposit date: | 2024-01-07 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural insights into the regulation of RyR1 by S100A1.
To Be Published
|
|
8VK4
 
 | |
1XQO
 
 | | Crystal structure of native Pa-AGOG, 8-oxoguanine DNA glycosylase from Pyrobaculum aerophilum | | Descriptor: | 8-oxoguanine DNA glycosylase | | Authors: | Lingaraju, G.M, Sartori, A.A, Kostrewa, D, Prota, A.E, Jiricny, J, Winkler, F.K. | | Deposit date: | 2004-10-13 | | Release date: | 2004-11-16 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | A DNA glycosylase from Pyrobaculum aerophilum with an 8-oxoguanine binding mode and a noncanonical helix-hairpin-helix structure
Structure, 13, 2005
|
|
