135D
 
 | |
1AN9
 
 | | D-AMINO ACID OXIDASE COMPLEX WITH O-AMINOBENZOATE | | Descriptor: | 2-AMINOBENZOIC ACID, D-AMINO ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Miura, R, Setoyama, C, Nishina, Y, Shiga, K, Mizutani, H, Miyahara, I, Hirotsu, K. | | Deposit date: | 1997-06-28 | | Release date: | 1997-11-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and mechanistic studies on D-amino acid oxidase x substrate complex: implications of the crystal structure of enzyme x substrate analog complex.
J.Biochem.(Tokyo), 122, 1997
|
|
6YUE
 
 | | Fragment of nitrate/nitrite sensor histidine kinase NarQ (R50S variant) | | Descriptor: | Nitrate/nitrite sensor protein NarQ | | Authors: | Gushchin, I, Melnikov, I, Polovinkin, V, Yuzhakova, A, Gordeliy, V. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sensor Histidine Kinase NarQ Activates via Helical Rotation, Diagonal Scissoring, and Eventually Piston-Like Shifts.
Int J Mol Sci, 21, 2020
|
|
1AMN
 
 | | TRANSITION STATE ANALOG: ACETYLCHOLINESTERASE COMPLEXED WITH M-(N,N,N-TRIMETHYLAMMONIO)TRIFLUOROACETOPHENONE | | Descriptor: | ACETYLCHOLINESTERASE, M-(N,N,N-TRIMETHYLAMMONIO)-2,2,2-TRIFLUORO-1,1-DIHYDROXYETHYLBENZENE, SULFATE ION | | Authors: | Harel, M, Silman, I, Sussman, J.L. | | Deposit date: | 1996-02-13 | | Release date: | 1996-04-03 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The X-ray structure of a transition state analog complex reveals the molecular origins of the catalytic power and substrate specificity of acetylcholinesterase.
J.Am.Chem.Soc., 118, 1996
|
|
5IZD
 
 | |
1A5Z
 
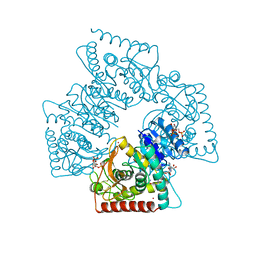 | | LACTATE DEHYDROGENASE FROM THERMOTOGA MARITIMA (TMLDH) | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, CADMIUM ION, L-LACTATE DEHYDROGENASE, ... | | Authors: | Auerbach, G, Ostendorp, R, Prade, L, Korndoerfer, I, Dams, T, Huber, R, Jaenicke, R. | | Deposit date: | 1998-02-18 | | Release date: | 1999-03-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Lactate dehydrogenase from the hyperthermophilic bacterium thermotoga maritima: the crystal structure at 2.1 A resolution reveals strategies for intrinsic protein stabilization.
Structure, 6, 1998
|
|
1AJT
 
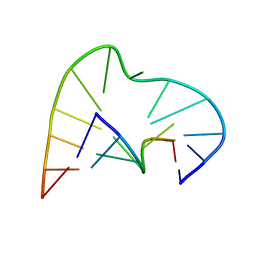 | | FIVE-NUCLEOTIDE BULGE LOOP FROM TETRAHYMENA THERMOPHILA GROUP I INTRON, NMR, 1 STRUCTURE | | Descriptor: | RNA (5'-R(*GP*AP*GP*UP*AP*CP*C)-3'), RNA (5'-R(*GP*GP*UP*AP*AP*UP*AP*AP*GP*CP*UP*C)-3') | | Authors: | Luebke, K.J, Landry, S.M, Tinoco Junior, I. | | Deposit date: | 1997-05-08 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of a five-nucleotide RNA bulge loop from a group I intron.
Biochemistry, 36, 1997
|
|
5OJW
 
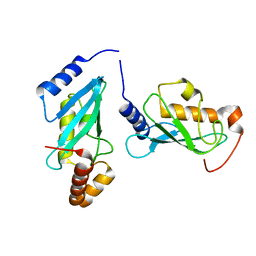 | | S. cerevisiae UBC13 - MMs2 complex | | Descriptor: | Ubiquitin-conjugating enzyme E2 13, Ubiquitin-conjugating enzyme variant MMS2 | | Authors: | Sharon, I, Rathi, R, Levin-Kravets, O, Attali, I, Prag, G. | | Deposit date: | 2017-07-24 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | S. cerevisiae UBC13 - MMs2 complex
To Be Published
|
|
1AJF
 
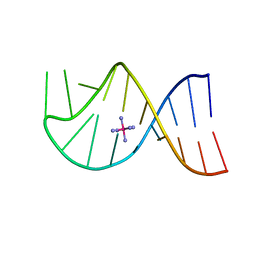 | |
3DWV
 
 | |
5J77
 
 | |
2EV2
 
 | | Structure of Rv1264N, the regulatory domain of the mycobacterial adenylyl cylcase Rv1264, at pH 8.5 | | Descriptor: | Hypothetical protein Rv1264/MT1302, OLEIC ACID | | Authors: | Findeisen, F, Tews, I, Sinning, I. | | Deposit date: | 2005-10-30 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structure of the regulatory domain of the adenylyl cyclase Rv1264 from Mycobacterium tuberculosis with bound oleic acid
J.Mol.Biol., 369, 2007
|
|
1A8Z
 
 | | STRUCTURE DETERMINATION OF A 16.8KDA COPPER PROTEIN RUSTICYANIN AT 2.1A RESOLUTION USING ANOMALOUS SCATTERING DATA WITH DIRECT METHODS | | Descriptor: | COPPER (I) ION, RUSTICYANIN | | Authors: | Harvey, I, Hao, Q, Duke, E.M.H, Ingledew, W.J, Hasnain, S.S. | | Deposit date: | 1998-03-30 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure determination of a 16.8 kDa copper protein at 2.1 A resolution using anomalous scattering data with direct methods.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
134D
 
 | |
1AA1
 
 | |
1BBP
 
 | | MOLECULAR STRUCTURE OF THE BILIN BINDING PROTEIN (BBP) FROM PIERIS BRASSICAE AFTER REFINEMENT AT 2.0 ANGSTROMS RESOLUTION. | | Descriptor: | BILIN BINDING PROTEIN, BILIVERDIN IX GAMMA CHROMOPHORE | | Authors: | Huber, R, Schneider, M, Mayr, I, Mueller, R, Deutzmann, R, Suter, F, Zuber, H, Falk, H, Kayser, H. | | Deposit date: | 1990-09-19 | | Release date: | 1991-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular structure of the bilin binding protein (BBP) from Pieris brassicae after refinement at 2.0 A resolution.
J.Mol.Biol., 198, 1987
|
|
1AQA
 
 | | SOLUTION STRUCTURE OF REDUCED MICROSOMAL RAT CYTOCHROME B5, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Banci, L, Bertini, I, Ferroni, F, Rosato, A. | | Deposit date: | 1997-07-28 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of reduced microsomal rat cytochrome b5.
Eur.J.Biochem., 249, 1997
|
|
5LKA
 
 | | Crystal structure of haloalkane dehalogenase LinB 140A+143L+177W+211L mutant (LinB86) from Sphingobium japonicum UT26 at 1.3 A resolution | | Descriptor: | Haloalkane dehalogenase, THIOCYANATE ION | | Authors: | Degtjarik, O, Rezacova, P, Iermak, I, Chaloupkova, R, Damborsky, J, Kuta Smatanova, I. | | Deposit date: | 2016-07-21 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.298 Å) | | Cite: | Engineering a de novo transport tunnel.
Acs Catalysis, 2016
|
|
1B41
 
 | | HUMAN ACETYLCHOLINESTERASE COMPLEXED WITH FASCICULIN-II, GLYCOSYLATED PROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Kryger, G, Harel, M, Shafferman, A, Silman, I, Sussman, J.L. | | Deposit date: | 1999-01-05 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structures of recombinant native and E202Q mutant human acetylcholinesterase complexed with the snake-venom toxin fasciculin-II.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
2CJA
 
 | | Crystal structure of Methanosarcina barkeri seryl-tRNA synthetase complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bilokapic, S, Maier, T, Ahel, D, Gruic-Sovulj, I, Soll, D, Weygand-Durasevic, I, Ban, N. | | Deposit date: | 2006-03-30 | | Release date: | 2006-06-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Unusual Seryl-tRNA Synthetase Reveals a Distinct Zinc-Dependent Mode of Substrate Recognition
Embo J., 25, 2006
|
|
3DEP
 
 | | Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43 | | Descriptor: | CHLORIDE ION, Signal recognition particle 43 kDa protein, YPGGSFDPLGLA | | Authors: | Holdermann, I, Stengel, K.F, Wild, K, Sinning, I. | | Deposit date: | 2008-06-10 | | Release date: | 2008-08-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43.
Science, 321, 2008
|
|
5S4J
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF054 | | Descriptor: | 6-chlorotetrazolo[1,5-b]pyridazine, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.124 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
2CIM
 
 | | Crystal structure of Methanosarcina barkeri seryl-tRNA synthetase | | Descriptor: | CHLORIDE ION, PLATINUM (II) ION, SERYL-TRNA SYNTHETASE, ... | | Authors: | Bilokapic, S, Maier, T, Ahel, D, Gruic-Sovulj, I, Soll, D, Weygand-Durasevic, I, Ban, N. | | Deposit date: | 2006-03-24 | | Release date: | 2006-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of the Unusual Seryl-tRNA Synthetase Reveals a Distinct Zinc-Dependent Mode of Substrate Recognition
Embo J., 25, 2006
|
|
2HRF
 
 | | Solution Structure of Cu(I) P174L HSco1 | | Descriptor: | COPPER (I) ION, SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Leontari, I, Martinelli, M, Palumaa, P, Sillard, R, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-07-20 | | Release date: | 2007-01-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Human Sco1 functional studies and pathological implications of the P174L mutant.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2HRN
 
 | | Solution Structure of Cu(I) P174L-HSco1 | | Descriptor: | COPPER (I) ION, SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Leontari, I, Martinelli, M, Palumaa, P, Sillard, R, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-07-20 | | Release date: | 2007-01-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Human Sco1 functional studies and pathological implications of the P174L mutant.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
