4NRA
 
 | | Crystal Structure of the bromodomain of human BAZ2B in complex with compound-6 E11322 | | Descriptor: | 1,2-ETHANEDIOL, 1-(8-chloro-1,3,4,5-tetrahydro-2H-pyrido[4,3-b]indol-2-yl)ethanone, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Chaikuad, A, Felletar, I, Ferguson, F.M, Filippakopoulos, P, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-11-26 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Targeting low-druggability bromodomains: fragment based screening and inhibitor design against the BAZ2B bromodomain.
J.Med.Chem., 56, 2013
|
|
4NNB
 
 | | Binary complex of ObcA with oxaloacetate | | Descriptor: | MAGNESIUM ION, OBCA, Oxalate Biosynthetic Component A, ... | | Authors: | Oh, J.T, Goo, E, Hwang, I, Rhee, S. | | Deposit date: | 2013-11-17 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Bacterial Quorum Sensing-mediated Oxalogenesis.
J.Biol.Chem., 289, 2014
|
|
4NR7
 
 | | Crystal structure of the bromodomain of human CREBBP in complex with an isoxazolyl-benzimidazole ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(3-chloro-4-methoxyphenyl)ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(2S)-2-(morpholin-4-yl)propyl]-1H-benzimidazole, CREB-binding protein | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Hay, D, Fedorov, O, Martin, S, Krojer, T, Nowak, R, von Delft, F, Brennan, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-11-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the bromodomain of human CREBBP in complex with an isoxazolyl-benzimidazole ligand
To be Published
|
|
4BX4
 
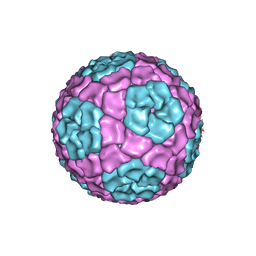 | | Fitting of the bacteriophage Phi8 P1 capsid protein into cryo-EM density | | Descriptor: | P1 | | Authors: | El Omari, K, Sutton, G, Ravantti, J.J, Zhang, H, Walter, T.S, Grimes, J.M, Bamford, D.H, Stuart, D.I, Mancini, E.J. | | Deposit date: | 2013-07-08 | | Release date: | 2013-08-07 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Plate Tectonics of Virus Shell Assembly and Reorganization in Phage Phi8, a Distant Relative of Mammalian Reoviruses
Structure, 21, 2013
|
|
4NLI
 
 | | Crystal structure of sheep beta-lactoglobulin (space group P3121) | | Descriptor: | Beta-lactoglobulin-1/B, SULFATE ION | | Authors: | Loch, J.I, Molenda, M, Kopec, M, Swiatek, S, Lewinski, K. | | Deposit date: | 2013-11-14 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of two crystal forms of sheep beta-lactoglobulin with EF-loop in closed conformation
Biopolymers, 101, 2014
|
|
5VRT
 
 | | Nonheme Iron Replacement in a Biosynthetic Nitric Oxide Reductase Model Performing O2 Reduction to Water: Co-bound FeBMb | | Descriptor: | COBALT (II) ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Reed, J, Shi, Y, Zhu, Q, Chakraborty, S, Mirs, E.N, Petrik, I.D, Bhagi-Damodaran, A, Ross, M, Moenne-Loccoz, P, Zhang, Y, Lu, Y. | | Deposit date: | 2017-05-11 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Manganese and Cobalt in the Nonheme-Metal-Binding Site of a Biosynthetic Model of Heme-Copper Oxidase Superfamily Confer Oxidase Activity through Redox-Inactive Mechanism.
J. Am. Chem. Soc., 139, 2017
|
|
4NV7
 
 | | Crystal Structure of Mesorhizobium Loti Arylamine N-acetyltransferase 1 In Complex With CoA | | Descriptor: | Arylamine N-acetyltransferase, COENZYME A | | Authors: | Xu, X.M, Haouz, A, Weber, P, Li de la sierra-gallay, I, Kubiak, X, Dupret, J.-M, Rodrigues-lima, F. | | Deposit date: | 2013-12-05 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Insight into cofactor recognition in arylamine N-acetyltransferase enzymes: structure of Mesorhizobium loti arylamine N-acetyltransferase in complex with coenzyme A.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4BG0
 
 | | Crystal structure of complement factors H and FHL-1 binding protein BBH06 or CRASP-2 from Borrelia burgdorferi | | Descriptor: | COMPLEMENT REGULATOR-ACQUIRING SURFACE PROTEIN 2 (CRASP-2 (CRASP-2) | | Authors: | Brangulis, K, Petrovskis, I, Baumanis, V, Tars, K. | | Deposit date: | 2013-03-22 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of Cspz, a Complement Regulator Factor H and Fhl-1 Binding Protein from Borrelia Burgdorferi.
FEBS J., 281, 2014
|
|
4AQI
 
 | | Structure of human S100A15 bound to zinc and calcium | | Descriptor: | CALCIUM ION, CHLORIDE ION, PROTEIN S100-A7A, ... | | Authors: | Murray, J.I, Tonkin, M.L, Whiting, A.L, Peng, F, Farnell, B, Hof, F, Boulanger, M.J. | | Deposit date: | 2012-04-17 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Characterization of S100A15 Reveals a Novel Zinc Coordination Site Among S100 Proteins and Altered Surface Chemistry with Functional Implications for Receptor Binding.
Bmc Struct.Biol., 12, 2012
|
|
4NUJ
 
 | |
4BJJ
 
 | | Sfc1-Sfc7 dimerization module | | Descriptor: | MERCURY (II) ION, TRANSCRIPTION FACTOR TAU SUBUNIT SFC1, TRANSCRIPTION FACTOR TAU SUBUNIT SFC7 | | Authors: | Taylor, N.M.I, Baudin, F, von Scheven, G, Muller, C.W. | | Deposit date: | 2013-04-18 | | Release date: | 2013-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | RNA Polymerase III-Specific General Transcription Factor Iiic Contains a Heterodimer Resembling Tfiif RAP30/RAP74.
Nucleic Acids Res., 41, 2013
|
|
4AK0
 
 | | Ligand controlled assembly of hexamers, dihexamers, and linear multihexamer structures by an engineered acylated insulin | | Descriptor: | INSULIN A CHAIN, INSULIN B CHAIN | | Authors: | Steensgaard, D.B, Schluckebier, G, Strauss, H.M, Norrman, M, Thomsen, J.K, Friderichsen, A.V, Havelund, S, Jonassen, I. | | Deposit date: | 2012-02-21 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Ligand Controlled Assembly of Hexamers, Dihexamers, and Linear Multihexamer Structures by the Engineered Acylated Insulin Degludec.
Biochemistry, 52, 2013
|
|
4NU9
 
 | | 2.30 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus with BME-free Cys289 | | Descriptor: | Betaine aldehyde dehydrogenase, SODIUM ION | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Stam, J, Shuvalova, L, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-03 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5VR3
 
 | | Crystal structure of the BRS domain of BRAF | | Descriptor: | BRAF, SULFATE ION | | Authors: | Thevakumaran, N, Maisonneuve, P, Kurinov, I, Lavoie, H, Marullo, S.A, Sahmi, M, Jin, T, Therrien, M, Sicheri, F. | | Deposit date: | 2017-05-10 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | MEK drives BRAF activation through allosteric control of KSR proteins.
Nature, 554, 2018
|
|
3SC7
 
 | | First crystal structure of an endo-inulinase, from Aspergillus ficuum: structural analysis and comparison with other GH32 enzymes. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Inulinase, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose, ... | | Authors: | Housen, I, Pouyez, J, Roussel, G, Mayard, A, Vandamme, A.M, Wouters, J, Michaux, C. | | Deposit date: | 2011-06-07 | | Release date: | 2012-06-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | First crystal structure of an endo-inulinase, INU2, from Aspergillus ficuum: Discovery of an extra-pocket in the catalytic domain responsible for its endo-activity.
Biochimie, 94, 2012
|
|
4NXY
 
 | | Crystal Structure of the GNAT domain of S. lividans PAT | | Descriptor: | Acyl-CoA synthetase, GLYCEROL, TRIFLUOROETHANOL | | Authors: | Rank, K.C, Tucker, A.C, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2013-12-09 | | Release date: | 2014-11-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Insights into the specificity of lysine acetyltransferases.
J.Biol.Chem., 289, 2014
|
|
5W2F
 
 | | Crystal Structure of the C-terminal Domain of Human eIF2D at 1.4 A resolution | | Descriptor: | Eukaryotic translation initiation factor 2D, FORMIC ACID | | Authors: | Vaidya, A.T, Lomakin, I.B, Joseph, N.N, Dmitriev, S.E, Steitz, T.A. | | Deposit date: | 2017-06-06 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of the C-terminal Domain of Human eIF2D and Its Implications on Eukaryotic Translation Initiation.
J. Mol. Biol., 429, 2017
|
|
5W1N
 
 | |
4O1Y
 
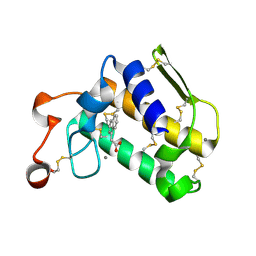 | | Crystal structure of Porcine Pancreatic Phospholipase A2 in complex with 1-Naphthaleneacetic acid | | Descriptor: | CALCIUM ION, NAPHTHALEN-1-YL-ACETIC ACID, Phospholipase A2, ... | | Authors: | Dileep, K.V, Remya, C, Tintu, I, Mandal, P.K, Karthe, P, Haridas, M, Sadasivan, C. | | Deposit date: | 2013-12-16 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Porcine Pancreatic Phospholipase A2 in complex with 1-Naphthaleneacetic acid
To be published
|
|
4F1O
 
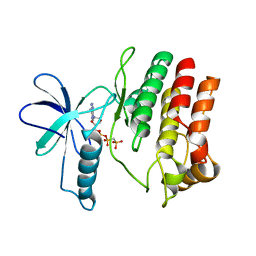 | | Crystal Structure of the L1180T mutant Roco4 Kinase Domain from D. discoideum bound to AppCp | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Serine/threonine-protein kinase roco4 | | Authors: | Gilsbach, B.K, Vetter, I.R, Wittinghofer, A, Kortholt, A. | | Deposit date: | 2012-05-07 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Roco kinase structures give insights into the mechanism of Parkinson disease-related leucine-rich-repeat kinase 2 mutations.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4F1T
 
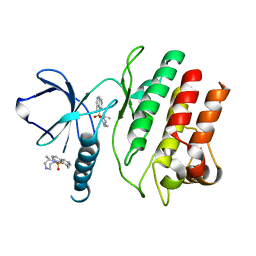 | | Crystal Structure of the Roco4 Kinase Domain from D. discoideum bound to the ROCK Inhibitor H1152 | | Descriptor: | (S)-2-METHYL-1-[(4-METHYL-5-ISOQUINOLINE)SULFONYL]-HOMOPIPERAZINE, Serine/threonine-protein kinase roco4 | | Authors: | Gilsbach, B.K, Vetter, I.R, Wittinghofer, A, Kortholt, A. | | Deposit date: | 2012-05-07 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Roco kinase structures give insights into the mechanism of Parkinson disease-related leucine-rich-repeat kinase 2 mutations.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FH3
 
 | |
3TS9
 
 | | Crystal Structure of the MDA5 Helicase Insert Domain | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, SULFATE ION | | Authors: | Berke, I.C, Modis, Y. | | Deposit date: | 2011-09-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | MDA5 cooperatively forms dimers and ATP-sensitive filaments upon binding double-stranded RNA.
Embo J., 31, 2012
|
|
4FCU
 
 | | 1.9 Angstrom Crystal Structure of 3-deoxy-manno-octulosonate Cytidylyltransferase (kdsB) from Acinetobacter baumannii without His-Tag Bound to the Active Site | | Descriptor: | 3-deoxy-manno-octulosonate cytidylyltransferase | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-25 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Crystal Structure of 3-deoxy-manno-octulosonate Cytidylyltransferase (kdsB) from Acinetobacter baumannii without His-Tag Bound to the Active Site.
TO BE PUBLISHED
|
|
4EYR
 
 | | Crystal structure of multidrug-resistant clinical isolate 769 HIV-1 protease in complex with ritonavir | | Descriptor: | HIV-1 PROTEASE, RITONAVIR | | Authors: | Liu, Z, Yedidi, R.S, Wang, Y, Brunzelle, J.S, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2012-05-01 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the mechanism of drug resistance: X-ray structure analysis of multi-drug resistant HIV-1 protease ritonavir complex.
Biochem.Biophys.Res.Commun., 431, 2013
|
|
