3RQX
 
 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis in complex with P1,P4-Di(adenosine-5') tetraphosphate | | Descriptor: | ADP/ATP-DEPENDENT NAD(P)H-HYDRATE DEHYDRATASE, BIS(ADENOSINE)-5'-TETRAPHOSPHATE, CHLORIDE ION, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-28 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RRB
 
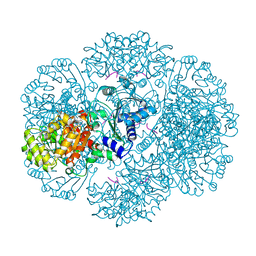 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Bifunctional NAD(P)H-hydrate repair enzyme Nnr, POTASSIUM ION, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-04-29 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
2R4S
 
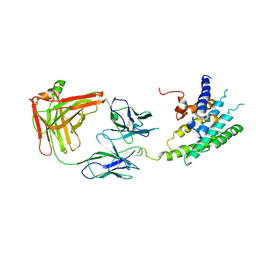 | | Crystal structure of the human beta2 adrenoceptor | | Descriptor: | Beta-2 adrenergic receptor, antibody for beta2 adrenoceptor, heavy chain, ... | | Authors: | Rasmussen, S.G.F, Choi, H.J, Rosenbaum, D.M, Kobilka, T.S, Thian, F.S, Edwards, P.C, Burghammer, M, Ratnala, V.R, Sanishvili, R, Fischetti, R.F, Schertler, G.F, Weis, W.I, Kobilka, B.K. | | Deposit date: | 2007-08-31 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of the human beta2 adrenergic G-protein-coupled receptor.
Nature, 450, 2007
|
|
3SJC
 
 | |
1SW7
 
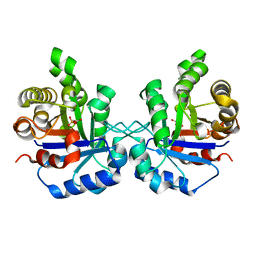 | | Triosephosphate isomerase from Gallus gallus, loop 6 mutant K174N, T175S, A176S | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-30 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
2QBL
 
 | | Crystal structure of ferric G248T cytochrome P450cam | | Descriptor: | CAMPHOR, Cytochrome P450-cam, POTASSIUM ION, ... | | Authors: | von Koenig, K, Makris, T.M, Sligar, S.D, Schlichting, I. | | Deposit date: | 2007-06-18 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Alteration of P450 Distal Pocket Solvent Leads to Impaired Proton Delivery and Changes in Heme Geometry.
Biochemistry, 46, 2007
|
|
4AHR
 
 | | Parallel screening of a low molecular weight compound library: do differences in methodology affect hit identification | | Descriptor: | 3-(1,3-benzodioxol-5-yl)propanoic acid, ACETIC ACID, GLYCEROL, ... | | Authors: | Wielens, J, Heady, S.J, Rhodes, D.I, Mulder, R.J, Dolezal, O, Deadman, J.J, Newman, J, Chalmers, D.K, Parker, M.W, Peat, T.S, Scanlon, M.J. | | Deposit date: | 2012-02-07 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Parallel Screening of Low Molecular Weight Fragment Libraries: Do Differences in Methodology Affect Hit Identification?
J.Biomol.Screen, 18, 2013
|
|
2VEL
 
 | | Structure-based enzyme engineering efforts with an inactive monomeric TIM variant: the importance of a single point mutation for generating an active site with suitable binding properties. | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, CHLORIDE ION, GLYCOSOMAL TRIOSEPHOSPHATE ISOMERASE | | Authors: | Alahuhta, M, Salin, M, Casteleijn, M.G, Kemmer, C, El-Sayed, I, Augustyns, K, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2007-10-24 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Protein Engineering Efforts with a Monomeric Tim Variant: The Importance of a Single Point Mutation for Generating an Active Site with Suitable Binding Properties.
Protein Eng.Des.Sel., 21, 2008
|
|
1SV1
 
 | |
2QBN
 
 | | Crystal structure of ferric G248V cytochrome P450cam | | Descriptor: | CAMPHOR, Cytochrome P450-cam, POTASSIUM ION, ... | | Authors: | von Koenig, K, Makris, T.M, Sligar, S.D, Schlichting, I. | | Deposit date: | 2007-06-18 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Alteration of P450 Distal Pocket Solvent Leads to Impaired Proton Delivery and Changes in Heme Geometry.
Biochemistry, 46, 2007
|
|
1T15
 
 | | Crystal Structure of the Brca1 BRCT Domains in Complex with the Phosphorylated Interacting Region from Bach1 Helicase | | Descriptor: | BRCA1 interacting protein C-terminal helicase 1, Breast cancer type 1 susceptibility protein | | Authors: | Clapperton, J.A, Manke, I.A, Lowery, D.M, Ho, T, Haire, L.F, Yaffe, M.B, Smerdon, S.J. | | Deposit date: | 2004-04-15 | | Release date: | 2004-05-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer
Nat.Struct.Mol.Biol., 11, 2004
|
|
4A8S
 
 | | Non-Catalytic Ions Direct the RNA-Dependent RNA Polymerase of Bacterial dsRNA virus phi6 from De Novo Initiation to Elongation | | Descriptor: | 5'-D(*TP*TP*TP*TP*CP*GP*CP*GP*TP*AP*GP*CP*GP)-3', ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wright, S, Poranen, M.M, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Noncatalytic Ions Direct the RNA-Dependent RNA Polymerase of Bacterial Double-Stranded RNA Virus Phi6 from De Novo Initiation to Elongation.
J.Virol., 86, 2012
|
|
1SZN
 
 | | THE STRUCTURE OF ALPHA-GALACTOSIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Golubev, A.M, Nagem, R.A.P, Brando Neto, J.R, Neustroev, K.N, Eneyskaya, E.V, Kulminskaya, A.A, Shabalin, K.A, Savel'ev, A.N, Polikarpov, I. | | Deposit date: | 2004-04-06 | | Release date: | 2004-08-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of alpha-galactosidase from Trichoderma reesei and its complex with galactose: implications for catalytic mechanism.
J.Mol.Biol., 339, 2004
|
|
3S3P
 
 | |
1T3P
 
 | | Half-sandwich arene ruthenium(II)-enzyme complex | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | McNae, I.W, Fishburne, K, Habtemariam, A, Hunter, T.M, Melchart, M, Wang, F, Walkinshaw, M.D, Sadler, P.J. | | Deposit date: | 2004-04-27 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Half-sandwich arene ruthenium(II)-enzyme complex
CHEM.COMMUN.(CAMB.), 16, 2004
|
|
1T0O
 
 | | The structure of alpha-galactosidase from Trichoderma reesei complexed with beta-D-galactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Golubev, A.M, Nagem, R.A.P, Brandao Neto, J.R, Neustroev, K.N, Eneyskaya, E.V, Kulminskaya, A.A, Shabalin, K.A, Savel'ev, A.N, Polikarpov, I. | | Deposit date: | 2004-04-12 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of alpha-galactosidase from Trichoderma reesei and its complex with galactose: implications for catalytic mechanism
J.Mol.Biol., 339, 2004
|
|
1T4G
 
 | | ATPase in complex with AMP-PNP | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wu, Y, He, Y, Moya, I.A, Qian, X, Luo, Y. | | Deposit date: | 2004-04-29 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Archaeal Recombinase RadA; A Snapshot of Its Extended Conformation.
Mol.Cell, 15, 2004
|
|
2QER
 
 | | Crystal structure of Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_1660 in the presence of dipeptide ala-pro | | Descriptor: | ALANINE, Cyclophilin-like protein, putative, ... | | Authors: | Wernimont, A.K, Lew, J, Hills, T, Hassanali, A, Lin, L, Wasney, G, Zhao, Y, Kozieradzki, I, Vedadi, M, Schapira, M, Bochkarev, A, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-26 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_1660 in the presence of dipeptide ala-pro.
To be Published
|
|
4ADM
 
 | | Crystal structure of Rv1098c in complex with meso-tartrate | | Descriptor: | FUMARATE HYDRATASE CLASS II, GLYCEROL, S,R MESO-TARTARIC ACID | | Authors: | Mechaly, A.E, Haouz, A, Miras, I, Weber, P, Shepard, W, Cole, S, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2011-12-27 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Conformational Changes Upon Ligand Binding in the Essential Class II Fumarase Rv1098C from Mycobacterium Tuberculosis.
FEBS Lett., 586, 2012
|
|
1T3K
 
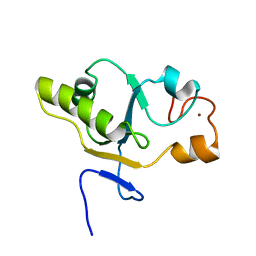 | | NMR structure of a CDC25-like dual-specificity tyrosine phosphatase of Arabidopsis thaliana | | Descriptor: | Dual-specificity tyrosine phosphatase, ZINC ION | | Authors: | Landrieu, I, da Costa, M, De Veylder, L, Dewitte, F, Vandepoele, K, Hassan, S, Wieruszeski, J.M, Faure, J.D, Inze, D, Lippens, G. | | Deposit date: | 2004-04-27 | | Release date: | 2004-09-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A small CDC25 dual-specificity tyrosine-phosphatase isoform in Arabidopsis thaliana.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
2QLK
 
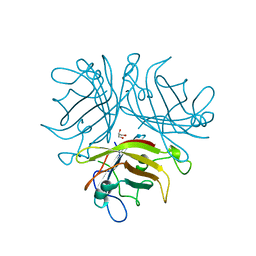 | | Adenovirus AD35 fibre head | | Descriptor: | Fiber, GLYCEROL | | Authors: | Liaw, Y.-C, Amiraslanov, I, Wang, H, Lieber, A. | | Deposit date: | 2007-07-13 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Identification of CD46 binding sites within the adenovirus serotype 35 fiber knob
J.Virol., 81, 2007
|
|
1T4Y
 
 | |
4AIZ
 
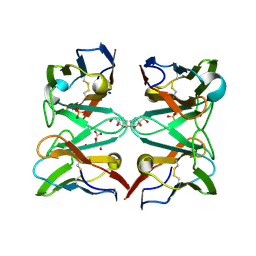 | | Crystallographic structure of 3mJL2 from the germinal line lambda 3 | | Descriptor: | 1,5:6,10-dianhydro-3,4,7,8-tetradeoxy-2,9-bis-C-(hydroxymethyl)-L-manno-decitol, CITRIC ACID, SULFATE ION, ... | | Authors: | Villalba, M.I, Luna, O.D, Rudino-Pinera, E, Sanchez, R, Sanchez-Lopez, R, Rojas-Trejo, S, Olamendi-Portugal, T, Fernandez-Velasco, D.A, Becerril, B. | | Deposit date: | 2012-02-15 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Site-Directed Mutagenesis Reveals Regions Implicated in the Stability and Fiber Formation of Human Lambda3R Light Chains.
J.Biol.Chem., 290, 2015
|
|
1T5Q
 
 | | Solution Structure of GIP(1-30)amide in TFE/Water | | Descriptor: | Gastric inhibitory polypeptide | | Authors: | Alana, I, Hewage, C.M, Malthouse, J.P.G, Parker, J.C, Gault, V.A, O'Harte, F.P.M. | | Deposit date: | 2004-05-05 | | Release date: | 2004-11-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the glucose-dependent insulinotropic polypeptide fragment, GIP(1-30)amide.
Biochem.Biophys.Res.Commun., 325, 2004
|
|
4AJY
 
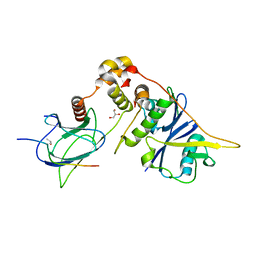 | | von Hippel-Lindau protein-ElonginB-ElonginC complex, bound to Hif1- alpha peptide | | Descriptor: | GLYCEROL, HYPOXIA-INDUCIBLE FACTOR 1-ALPHA, TRANSCRIPTION ELONGATION FACTOR B POLYPEPTIDE 1, ... | | Authors: | Van Molle, I, Thomann, A, Buckley, D.L, So, E.C, Lang, S, Crews, C.M, Ciulli, A. | | Deposit date: | 2012-02-21 | | Release date: | 2012-11-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Dissecting Fragment-Based Lead Discovery at the Von Hippel-Lindau Protein:Hypoxia Inducible Factor 1Alpha Protein-Protein Interface.
Chem.Biol., 19, 2012
|
|
