4H2U
 
 | | Crystal structure of Bradyrhizobium japonicum glycine:[carrier protein] ligase complexed with cognate carrier protein and ATP | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Luic, M, Weygand-Durasevic, I, Ivic, N, Mocibob, M. | | Deposit date: | 2012-09-13 | | Release date: | 2013-03-06 | | Last modified: | 2013-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Adaptation of aminoacyl-tRNA synthetase catalytic core to carrier protein aminoacylation.
Structure, 21, 2013
|
|
2WBG
 
 | | Structure of family 1 beta-glucosidase from Thermotoga maritima in complex with 3-imino-2-oxa-(+)-castanospermine | | Descriptor: | (3Z,5S,6R,7S,8R,8aR)-3-(octylimino)hexahydro[1,3]oxazolo[3,4-a]pyridine-5,6,7,8-tetrol, ACETATE ION, BETA-GLUCOSIDASE A | | Authors: | Aguilar, M, Gloster, T.M, Turkenburg, J.P, Garcia-Moreno, M.I, Ortiz Mellet, C, Davies, G.J, Garcia Fernandez, J.M. | | Deposit date: | 2009-02-27 | | Release date: | 2009-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Glycosidase Inhibition by Ring-Modified Castanospermine Analogues: Tackling Enzyme Selectivity by Inhibitor Tailoring.
Org.Biomol.Chem., 7, 2009
|
|
1E7K
 
 | | Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment | | Descriptor: | 15.5 KD RNA BINDING PROTEIN, RNA (5'-R(*GP*CP*CP*AP*AP*UP*GP*AP*GP*GP*UP*UP*UP* AP*UP*CP*CP*GP*AP*GP*G*C(-3') | | Authors: | Vidovic, I, Nottrott, S, Harthmuth, K, Luhrmann, R, Ficner, R. | | Deposit date: | 2000-08-29 | | Release date: | 2001-02-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Spliceosomal 15.5Kd Protein Bound to a U4 Snrna Fragment
Mol.Cell, 6, 2000
|
|
5A2K
 
 | | Crystal structure of scFv-SM3 in complex with APD-TGalNAc-RP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, ANTIGEN TN, ... | | Authors: | Martinez-Saez, N, Castro-Lopez, J, Valero-Gonzalez, J, Madariaga, D, Companon, I, Somovilla, V.J, Salvado, M, Asensio, J.L, Jimenez-Barbero, J, Avenoza, A, Busto, J.H, Bernardes, G.J.L, Peregrina, J.M, Hurtado-Guerrero, R, Corzana, F. | | Deposit date: | 2015-05-20 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Deciphering the Non-Equivalence of Serine and Threonine O-Glycosylation Points: Implications for Molecular Recognition of the Tn Antigen by an Anti-Muc1 Antibody.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7S4R
 
 | | Serial Macromolecular Crystallography at ALBA Synchrotron Light Source - alpha Spectrin-SH3 domain | | Descriptor: | Spectrin alpha chain, non-erythrocytic 1 | | Authors: | Martin-Garcia, J.M, Botha, S, Hu, H, Jernigan, R, Castellvi, A, Lisova, S, Gil, F, Calisto, B, Crespo, I, Roy-Chowdbury, S, Grieco, A, Ketawala, G, Weierstall, U, Spence, J, Fromme, P, Zatsepin, N, Boer, R, Carpena, X. | | Deposit date: | 2021-09-09 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Serial macromolecular crystallography at ALBA Synchrotron Light Source.
J.Synchrotron Radiat., 29, 2022
|
|
4L8Q
 
 | | Crystal structure of Canavalia grandiflora seed lectin complexed with X-Man. | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CADMIUM ION, CALCIUM ION, ... | | Authors: | Barroso-Neto, I.L, Rocha, B.A.M, Simoes, R.C, Bezerra, M.J.B, Pereira-Junior, F.N, Osterne, V.J.S, Nascimento, K.S, Nagano, C.S, Delatorre, P, Sampaio, A.H, Cavada, B.S. | | Deposit date: | 2013-06-17 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Vasorelaxant activity of Canavalia grandiflora seed lectin: A structural analysis.
Arch.Biochem.Biophys., 543, 2014
|
|
5EKH
 
 | |
5FGT
 
 | | Thaumatin solved by native sulphur-SAD using free-electron laser radiation | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K.J, Meinhart, A, Barends, T.R.M, Foucar, L, Gorel, A, Aquila, A, Botha, S, Doak, R.B, Koglin, J, Liang, M, Shoeman, R.L, Williams, G.K, Boutet, S, Schlichting, I. | | Deposit date: | 2015-12-21 | | Release date: | 2016-06-08 | | Last modified: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein structure determination by single-wavelength anomalous diffraction phasing of X-ray free-electron laser data.
Iucrj, 3, 2016
|
|
8JZD
 
 | | Crystal structure of Escherichia coli NarJ in complex with the signal peptide of E. coli NarG | | Descriptor: | Nitrate reductase molybdenum cofactor assembly chaperone NarJ, Respiratory nitrate reductase 1 alpha chain | | Authors: | Song, W.S, Kim, J.H, Namgung, B, Cho, H.Y, Oh, H.B, Yoon, S.I. | | Deposit date: | 2023-07-05 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Complementary hydrophobic interaction of the redox enzyme maturation protein NarJ with the signal peptide of the respiratory nitrate reductase NarG.
Int.J.Biol.Macromol., 262, 2024
|
|
5FGX
 
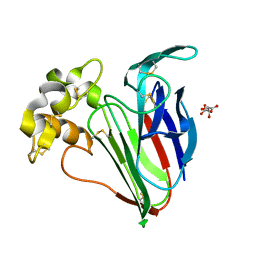 | | Thaumatin solved by native sulphur SAD using synchrotron radiation | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K.J, Meinhart, A, Barends, T.R.M, Foucar, L, Gorel, A, Aquila, A, Botha, S, Doak, R.B, Koglin, J, Liang, M, Shoeman, R.L, Williams, G.J, Boutet, S, Schlichting, I. | | Deposit date: | 2015-12-21 | | Release date: | 2016-06-08 | | Method: | X-RAY DIFFRACTION (2.134 Å) | | Cite: | Protein structure determination by single-wavelength anomalous diffraction phasing of X-ray free-electron laser data.
Iucrj, 3, 2016
|
|
6ZHI
 
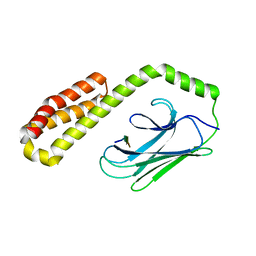 | |
3ZTC
 
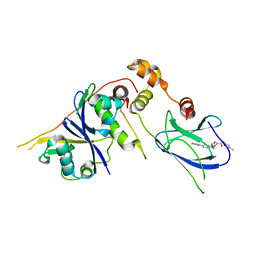 | | pVHL54-213-EloB-EloC complex _ (2S,4R)-N-((1,1'-biphenyl)-4-ylmethyl)- 4-hydroxy-1-(2-(3-methylisoxazol-5-yl)acetyl)pyrrolidine-2- carboxamide | | Descriptor: | (4R)-N-(BIPHENYL-4-YLMETHYL)-4-HYDROXY-1-[(3-METHYLISOXAZOL-5-YL)ACETYL]-L-PROLINAMIDE, TRANSCRIPTION ELONGATION FACTOR B POLYPEPTIDE 1, TRANSCRIPTION ELONGATION FACTOR B POLYPEPTIDE 2, ... | | Authors: | Van Molle, I, Buckley, D.L, Crews, C.M, Ciulli, A. | | Deposit date: | 2011-07-06 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Dissecting Fragment-Based Lead Discovery at the Von Hippel-Lindau Protein:Hypoxia Inducible Factor 1Alpha Protein-Protein Interface.
Chem.Biol., 19, 2012
|
|
5AKB
 
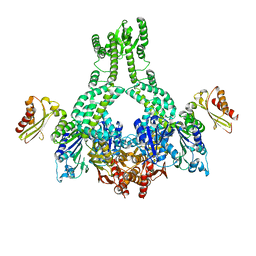 | | MutS in complex with the N-terminal domain of MutL - crystal form 1 | | Descriptor: | DNA MISMATCH REPAIR PROTEIN MUTL, DNA MISMATCH REPAIR PROTEIN MUTS, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Groothuizen, F.S, Winkler, I, Cristovao, M, Fish, A, Winterwerp, H.H.K, Reumer, A, Marx, A.D, Hermans, N, Nicholls, R.A, Murshudov, G.N, Lebbink, J.H.G, Friedhoff, P, Sixma, T.K. | | Deposit date: | 2015-03-03 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.71 Å) | | Cite: | MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA.
Elife, 4, 2015
|
|
6YZ7
 
 | | H11-D4, SARS-CoV-2 RBD, CR3022 ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Cr3022, Antibody light chain, ... | | Authors: | Naismith, J.H, Ren, J, Zhou, D, Zhao, Y, Stuart, D.I. | | Deposit date: | 2020-05-06 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural characterisation of a nanobody derived from a naive library that neutralises SARS-CoV-2
To Be Published, 2020
|
|
2VYU
 
 | | CRYSTAL STRUCTURE OF CHOLINE BINDING PROTEIN F FROM STREPTOCOCCUS PNEUMONIAE IN THE PRESENCE OF A PEPTIDOGLYCAN ANALOGUE (TETRASACCHARIDE-PENTAPEPTIDE) | | Descriptor: | CHOLINE BINDING PROTEIN F, CHOLINE ION | | Authors: | Perez-Dorado, I, Molina, R, Hermoso, J.A, Mobashery, S. | | Deposit date: | 2008-07-28 | | Release date: | 2009-02-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Cbpf, a Bifunctional Choline-Binding Protein and Autolysis Regulator from Streptococcus Pneumoniae.
Embo Rep., 10, 2009
|
|
4ZVA
 
 | |
2VWT
 
 | | Crystal structure of YfaU, a metal ion dependent class II aldolase from Escherichia coli K12 - Mg-pyruvate product complex | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Rea, D, Rakus, J.F, Gerlt, J.A, Fulop, V, Bugg, T.D.H, Roper, D.I. | | Deposit date: | 2008-06-26 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure and Functional Assignment of Yfau, a Metal Ion Dependent Class II Aldolase from Escherichia Coli K12.
Biochemistry, 47, 2008
|
|
6Z2M
 
 | | H11-D4, SARS-CoV-2 RBD, CR3022 ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CR3022 antibody, Spike glycoprotein, ... | | Authors: | Naismith, J.H, Ren, J, Zhou, D, Zhao, Y, Stuart, D.I. | | Deposit date: | 2020-05-17 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural characterisation of a nanobody derived from a naive library that neutralises SARS-CoV-2
To Be Published
|
|
2VZK
 
 | | Structure of the acyl-enzyme complex of an N-terminal nucleophile (Ntn) hydrolase, OAT2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLUTAMATE N-ACETYLTRANSFERASE 2 ALPHA CHAIN, ... | | Authors: | Iqbal, A, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2008-08-01 | | Release date: | 2008-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Anatomy of a Simple Acyl Intermediate in Enzyme Catalysis: Combined Biophysical and Modeling Studies on Ornithine Acetyl Transferase.
J.Am.Chem.Soc., 131, 2009
|
|
1D2I
 
 | | CRYSTAL STRUCTURE OF RESTRICTION ENDONUCLEASE BGLII COMPLEXED WITH DNA 16-MER | | Descriptor: | DNA (5'-D(*TP*AP*TP*TP*AP*TP*AP*GP*AP*TP*CP*TP*AP*TP*AP*A)-3'), MAGNESIUM ION, PROTEIN (RESTRICTION ENDONUCLEASE BGLII) | | Authors: | Lukacs, C.M, Kucera, R, Schildkraut, I, Aggarwal, A.K. | | Deposit date: | 1999-09-23 | | Release date: | 2000-02-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Understanding the immutability of restriction enzymes: crystal structure of BglII and its DNA substrate at 1.5 A resolution.
Nat.Struct.Biol., 7, 2000
|
|
7S0I
 
 | | CRYSTAL STRUCTURE OF N1 NEURAMINIDASE FROM A/Michigan/45/2015(H1N1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neuraminidase, ... | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2021-08-30 | | Release date: | 2021-12-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.892 Å) | | Cite: | A Novel Recombinant Influenza Virus Neuraminidase Vaccine Candidate Stabilized by a Measles Virus Phosphoprotein Tetramerization Domain Provides Robust Protection from Virus Challenge in the Mouse Model.
Mbio, 12, 2021
|
|
5IHU
 
 | | Crystal structure of bovine Fab B11 | | Descriptor: | bovine Fab B11 heavy chain, bovine Fab B11 light chain | | Authors: | Stanfield, R, Wilson, I. | | Deposit date: | 2016-02-29 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Conservation and diversity in the ultralong third heavy-chain complementarity-determining region of bovine antibodies.
Sci Immunol, 1, 2016
|
|
5A80
 
 | | Crystal structure of human JMJD2A in complex with compound 40 | | Descriptor: | 1,2-ETHANEDIOL, 2-[5-[2-(3-methoxyphenyl)ethanoylamino]-2-oxidanyl-phenyl]pyridine-4-carboxylic acid, LYSINE-SPECIFIC DEMETHYLASE 4A, ... | | Authors: | Nowak, R, Velupillai, S, Krojer, T, Gileadi, C, Johansson, C, Korczynska, M, Le, D.D, Younger, N, Gregori-Puigjane, E, Tumber, A, Iwasa, E, Pollock, S.B, Ortiz Torres, I, Kopec, J, Tallant, C, Froese, S, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Shoichet, B.K, Fujimori, D.G, Oppermann, U. | | Deposit date: | 2015-07-11 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Docking and Linking of Fragments to Discover Jumonji Histone Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
7S3F
 
 | | Structure of cofactor pyridoxal 5-phosphate bound human ornithine decarboxylase in complex with its inhibitor 1-amino-oxy-3-aminopropane | | Descriptor: | 3-AMINOOXY-1-AMINOPROPANE, Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zhou, X.E, Suino-Powell, K, Schultz, C.R, Aleiwi, B, Brunzelle, J.S, Lamp, J, Vega, I.E, Ellsworth, E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2021-09-06 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis of binding and inhibition of ornithine decarboxylase by 1-amino-oxy-3-aminopropane.
Biochem.J., 478, 2021
|
|
3LDM
 
 | |
