1HNG
 
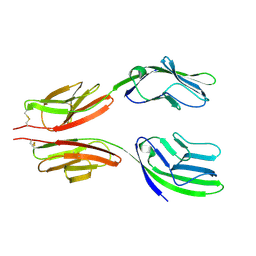 | | CRYSTAL STRUCTURE AT 2.8 ANGSTROMS RESOLUTION OF A SOLUBLE FORM OF THE CELL ADHESION MOLECULE CD2 | | Descriptor: | CD2 | | Authors: | Jones, E.Y, Davis, S.J, Williams, A.F, Harlos, K, Stuart, D.I. | | Deposit date: | 1994-08-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure at 2.8 A resolution of a soluble form of the cell adhesion molecule CD2.
Nature, 360, 1992
|
|
5MJQ
 
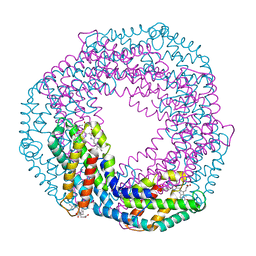 | | Single-shot pink beam serial crystallography: Phycocyanin (One chip) | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Meents, A, Oberthuer, D, Lieske, J, Srajer, V, Sarrou, I. | | Deposit date: | 2016-12-01 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Pink-beam serial crystallography.
Nat Commun, 8, 2017
|
|
6EFV
 
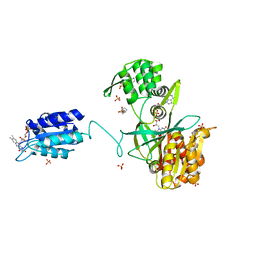 | | The NADPH-dependent sulfite reductase flavoprotein adopts an extended conformation that is unique to this diflavin reductase | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tavolieri, A.M, Askenasy, I, Murray, D.T, Pennington, J.M, Stroupe, M.E. | | Deposit date: | 2018-08-17 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | NADPH-dependent sulfite reductase flavoprotein adopts an extended conformation unique to this diflavin reductase.
J. Struct. Biol., 205, 2019
|
|
8E01
 
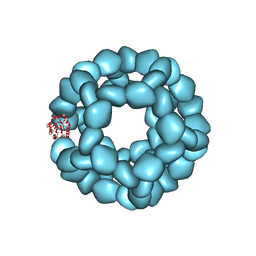 | | Structure of engineered nano-cage fusion protein | | Descriptor: | 2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase | | Authors: | Moustafa, I.M, Hafenstein, S.L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-11-16 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Intranasal SARS-CoV-2 RBD decorated nanoparticle vaccine enhances viral clearance in the Syrian hamster model.
Biorxiv, 2022
|
|
1MR7
 
 | | Crystal Structure of Streptogramin A Acetyltransferase | | Descriptor: | Streptogramin A Acetyltransferase | | Authors: | Kehoe, L.E, Snidwongse, J, Courvalin, P, Rafferty, J.B, Murray, I.A. | | Deposit date: | 2002-09-18 | | Release date: | 2003-08-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Synercid (Quinupristin-Dalfopristin) Resistance in Gram-positive Bacterial Pathogens
J.Biol.Chem., 278, 2003
|
|
6LCD
 
 | |
8AYZ
 
 | | Poliovirus type 2 (strain MEF-1) virus-like particle in complex with capsid binder compound 17 | | Descriptor: | 4-[[4-[1,3-bis(oxidanylidene)isoindol-2-yl]phenyl]sulfonylamino]benzoic acid, Capsid protein, VP0, ... | | Authors: | Bahar, M.W, Fry, E.E, Stuart, D.I. | | Deposit date: | 2022-09-04 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (1.88 Å) | | Cite: | A conserved glutathione binding site in poliovirus is a target for antivirals and vaccine stabilisation.
Commun Biol, 5, 2022
|
|
8AYY
 
 | | Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8) in complex with GSH and Pleconaril | | Descriptor: | 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-TRIFLUOROMETHYL-[1,2,4]OXADIAZOLE, Capsid protein, VP0, ... | | Authors: | Bahar, M.W, Fry, E.E, Stuart, D.I. | | Deposit date: | 2022-09-04 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A conserved glutathione binding site in poliovirus is a target for antivirals and vaccine stabilisation.
Commun Biol, 5, 2022
|
|
8AYX
 
 | | Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8) in complex with GSH and GPP3 | | Descriptor: | 1-[(3S)-5-[4-[(E)-ETHOXYIMINOMETHYL]PHENOXY]-3-METHYL-PENTYL]-3-PYRIDIN-4-YL-IMIDAZOLIDIN-2-ONE, Capsid protein, VP0, ... | | Authors: | Bahar, M.W, Fry, E.E, Stuart, D.I. | | Deposit date: | 2022-09-04 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A conserved glutathione binding site in poliovirus is a target for antivirals and vaccine stabilisation.
Commun Biol, 5, 2022
|
|
6EMF
 
 | |
6RTR
 
 | | Piperideine-6-carboxylate dehydrogenase from Streptomyces clavuligerus | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Hasse, D, Huelsemann, J, Carlsson, G, Andersson, I. | | Deposit date: | 2019-05-26 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and mechanism of piperideine-6-carboxylate dehydrogenase from Streptomyces clavuligerus.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1GUM
 
 | | HUMAN GLUTATHIONE TRANSFERASE A4-4 WITHOUT LIGANDS | | Descriptor: | PROTEIN (GLUTATHIONE TRANSFERASE A4-4) | | Authors: | Bruns, C.M, Hubatsch, I, Ridderstrom, M, Mannervik, B, Tainer, J.A. | | Deposit date: | 1998-06-11 | | Release date: | 1999-01-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human glutathione transferase A4-4 crystal structures and mutagenesis reveal the basis of high catalytic efficiency with toxic lipid peroxidation products
J.Mol.Biol., 288, 1999
|
|
6S7C
 
 | | Crystal structure of CARM1 in complex with inhibitor UM079 | | Descriptor: | 1-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(3-azanylpropyl)amino]propyl]guanidine, Histone-arginine methyltransferase CARM1 | | Authors: | Gunnell, E.A, Muhsen, U, Dowden, J, Dreveny, I. | | Deposit date: | 2019-07-04 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical evaluation of bisubstrate inhibitors of protein arginine N-methyltransferases PRMT1 and CARM1 (PRMT4).
Biochem.J., 477, 2020
|
|
5M8J
 
 | | Crystal structure of Eremococcus coleocola manganese transporter mutant H236A | | Descriptor: | Divalent metal cation transporter MntH | | Authors: | Manatschal, C, Ehrnstorfer, I.A, Arnold, F.M, Laederach, J, Dutzler, R. | | Deposit date: | 2016-10-29 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural and mechanistic basis of proton-coupled metal ion transport in the SLC11/NRAMP family.
Nat Commun, 8, 2017
|
|
6RF4
 
 | | Crystal structure of the potassium-pumping S254A mutant of the light-driven sodium pump KR2 in the pentameric form, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
5M95
 
 | | STAPHYLOCOCCUS CAPITIS DIVALENT METAL ION TRANSPORTER (DMT) IN COMPLEX WITH MANGANESE | | Descriptor: | CAMELID ANTIBODY FRAGMENT, NANOBODY, Divalent metal cation transporter MntH, ... | | Authors: | Ehrnstorfer, I.A, Geertsma, E.R, Pardon, E, Steyaert, J, Dutzler, R. | | Deposit date: | 2016-10-31 | | Release date: | 2016-11-30 | | Last modified: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure Of A Slc11 (Nramp) Transporter Reveals The Basis For Transition-Metal Ion Transport.
Nat.Struct.Mol.Biol., 21, 2014
|
|
6RF3
 
 | | Crystal structure of the potassium-pumping G263F mutant of the light-driven sodium pump KR2 in the pentameric form, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, SODIUM ION, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
3GHP
 
 | |
4Y1K
 
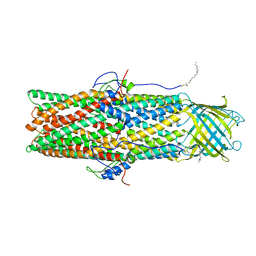 | |
4IV3
 
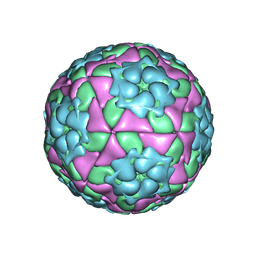 | | Crystal structure of recombinant foot-and-mouth-disease virus A22-H2093C empty capsid | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Porta, C, Kotecha, A, Burman, A, Jackson, T, Ren, J, Loureiro, S, Jones, I.M, Fry, E.E, Stuart, D.I, Charleston, B. | | Deposit date: | 2013-01-22 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Rational engineering of recombinant picornavirus capsids to produce safe, protective vaccine antigen.
Plos Pathog., 9, 2013
|
|
6EDF
 
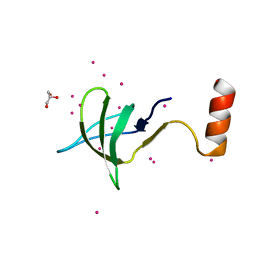 | | Fragment of a tyrosine-protein kinase | | Descriptor: | FYN, GLYCEROL, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Huang, H, Sochirca, I, Liu, K, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Sidhu, S.S, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-08-09 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Fragment of a tyrosine-protein kinase
To be Published
|
|
4O9K
 
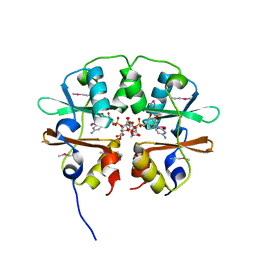 | | Crystal structure of the CBS pair of a putative D-arabinose 5-phosphate isomerase from Methylococcus capsulatus in complex with CMP-Kdo | | Descriptor: | Arabinose 5-phosphate isomerase, CYTIDINE 5'-MONOPHOSPHATE 3-DEOXY-BETA-D-GULO-OCT-2-ULO-PYRANOSONIC ACID, GLYCEROL | | Authors: | Shabalin, I.G, Cooper, D.R, Shumilin, I.A, Zimmerman, M.D, Majorek, K.A, Hammonds, J, Hillerich, B.S, Nawar, A, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-01-02 | | Release date: | 2014-01-22 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and kinetic properties of D-arabinose 5-phosphate isomerase from Methylococcus capsulatus
To be Published
|
|
4XTO
 
 | | Crystal structure of the light-driven sodium pump KR2 in the pentameric red form, pH 5.6 | | Descriptor: | EICOSANE, SODIUM ION, Sodium pumping rhodopsin | | Authors: | Gushchin, I, Shevchenko, V, Polovinkin, V, Gordeliy, V. | | Deposit date: | 2015-01-23 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a light-driven sodium pump.
Nat.Struct.Mol.Biol., 22, 2015
|
|
6E9P
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - open form with BRD0059 bound | | Descriptor: | (2R,3S,4R)-3-(2',6'-difluoro-4'-methyl[1,1'-biphenyl]-4-yl)-4-(fluoromethyl)azetidine-2-carbonitrile, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Chang, C, Michalska, K, Maltseva, N.I, Jedrzejczak, R, McCarren, P, Nag, P.P, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-08-01 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.569 Å) | | Cite: | Crystal structure of tryptophan synthase from M. tuberculosis - closed form with BRD6309 bound
To be Published
|
|
6KVA
 
 | | Structure of anti-hCXCR2 abN48-2 in complex with its CXCR2 epitope | | Descriptor: | 1,2-ETHANEDIOL, Peptide from C-X-C chemokine receptor type 2, heavy chain, ... | | Authors: | Xiang, J.C, Yan, L, Yang, B, Wilson, I.A. | | Deposit date: | 2019-09-03 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selection of a picomolar antibody that targets CXCR2-mediated neutrophil activation and alleviates EAE symptoms.
Nat Commun, 12, 2021
|
|
