2F95
 
 | | M intermediate structure of sensory rhodopsin II/transducer complex in combination with the ground state structure | | Descriptor: | RETINAL, Sensory rhodopsin II, Sensory rhodopsin II transducer, ... | | Authors: | Moukhametzianov, R.I, Klare, J.P, Efremov, R.G, Baecken, C, Goeppner, A, Labahn, J, Engelhard, M, Bueldt, G, Gordeliy, V.I. | | Deposit date: | 2005-12-05 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of the signal in sensory rhodopsin and its transfer to the cognate transducer.
Nature, 440, 2006
|
|
3R5L
 
 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Deazaflavin-dependent nitroreductase | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayyar, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-18 | | Release date: | 2012-01-18 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
8S0N
 
 | |
135D
 
 | |
8SFN
 
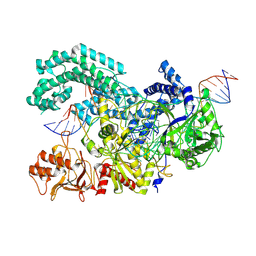 | |
8SFI
 
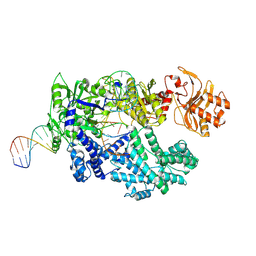 | | WT CRISPR-Cas12a with a 8bp R-loop | | Descriptor: | CRISPR-associated endonuclease Cas12a, DNA (5'-D(P*CP*TP*TP*AP*TP*CP*AP*CP*TP*AP*AP*AP*AP*GP*AP*TP*CP*GP*GP*AP*AP*G)-3'), DNA (5'-D(P*CP*TP*TP*CP*CP*GP*AP*TP*CP*TP*TP*TP*TP*AP*GP*TP*GP*AP*TP*A)-3'), ... | | Authors: | Strohkendl, I, Taylor, D.W. | | Deposit date: | 2023-04-11 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | WT CRISPR-Cas12a with a 8bp R-loop
To Be Published
|
|
3RO7
 
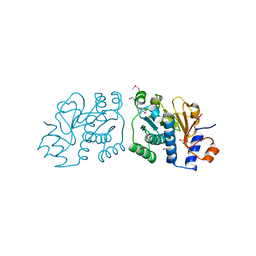 | | Crystal Structure of Mouse Apolipoprotein A-I Binding Protein in Complex with Thymine. | | Descriptor: | Apolipoprotein A-I-binding protein, SULFATE ION, THYMINE | | Authors: | Shumilin, I.A, Jha, K.N, Cymborowski, M, Herr, J.C, Minor, W. | | Deposit date: | 2011-04-25 | | Release date: | 2012-07-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
5HCQ
 
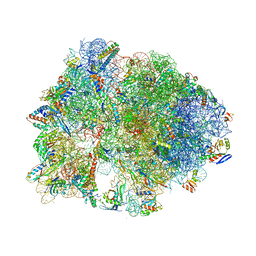 | | Crystal structure of antimicrobial peptide Oncocin d15-19 bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Gagnon, M.G, Roy, R.N, Lomakin, I.B, Florin, T, Mankin, A.S, Steitz, T.A. | | Deposit date: | 2016-01-04 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structures of proline-rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition.
Nucleic Acids Res., 44, 2016
|
|
4MHI
 
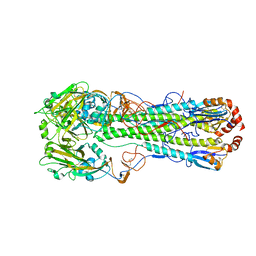 | | Crystal structure of a H5N1 influenza virus hemagglutinin from A/goose/Guangdong/1/96 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2013-08-29 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | A Unique and Conserved Neutralization Epitope in H5N1 Influenza Viruses Identified by an Antibody against the A/Goose/Guangdong/1/96 Hemagglutinin.
J.Virol., 87, 2013
|
|
8SFQ
 
 | |
4KAS
 
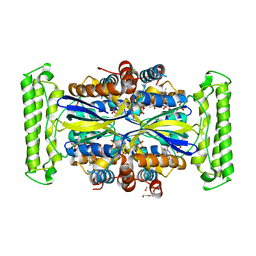 | | Crystal structure of FDTS from T. maritima mutant (H53D) with FAD and dUMP | | Descriptor: | 2'-DEOXYURIDINE-5'-MONOPHOSPHATE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mathews, I.I. | | Deposit date: | 2013-04-22 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Flavin-Dependent Thymidylate Synthase as a Drug Target for Deadly Microbes: Mutational Study and a Strategy for Inhibitor Design.
J Bioterror Biodef, Suppl 12, 2013
|
|
8SFO
 
 | |
1AMQ
 
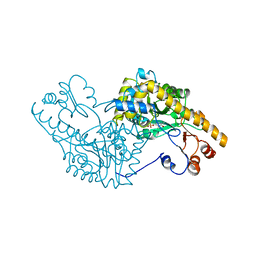 | | X-RAY CRYSTALLOGRAPHIC STUDY OF PYRIDOXAMINE 5'-PHOSPHATE-TYPE ASPARTATE AMINOTRANSFERASES FROM ESCHERICHIA COLI IN THREE FORMS | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE | | Authors: | Miyahara, I, Hirotsu, K, Hayashi, H, Kagamiyama, H. | | Deposit date: | 1994-07-01 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystallographic study of pyridoxamine 5'-phosphate-type aspartate aminotransferases from Escherichia coli in three forms.
J.Biochem.(Tokyo), 116, 1994
|
|
3GG6
 
 | | Crystal structure of the NUDIX domain of human NUDT18 | | Descriptor: | Nucleoside diphosphate-linked moiety X motif 18 | | Authors: | Tresaugues, L, Siponen, M.I, Lehtio, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the NUDIX domain of human NUDT18
To be Published
|
|
1AO2
 
 | | cobalt(III)-deglycopepleomycin determined by NMR studies | | Descriptor: | AGLYCON OF PEPLOMYCIN, COBALT (III) ION, HYDROGEN PEROXIDE | | Authors: | Caceres-Cortes, J, Sugiyama, H, Ikudome, K, Saito, I, Wang, A.H.-J. | | Deposit date: | 1997-07-16 | | Release date: | 1999-07-30 | | Last modified: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | Structures of cobalt(III)-pepleomycin and cobalt(III)-deglycopepleomycin (green forms) determined by NMR studies.
Eur.J.Biochem., 244, 1997
|
|
8SFR
 
 | | WT CRISPR-Cas12a post nontarget strand cleavage. | | Descriptor: | CRISPR-associated endonuclease Cas12a, DNA (34-MER), DNA (5'-D(P*CP*TP*TP*CP*CP*GP*AP*TP*CP*TP*TP*TP*TP*AP*GP*TP*GP*AP*T)-3'), ... | | Authors: | Strohkendl, I, Taylor, D.W. | | Deposit date: | 2023-04-11 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | WT CRISPR-Cas12a post nontarget strand cleavage.
To Be Published
|
|
2F80
 
 | | HIV-1 Protease mutant D30N complexed with inhibitor TMC114 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, POL POLYPROTEIN, ... | | Authors: | Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2005-12-01 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Effectiveness of Nonpeptide Clinical Inhibitor TMC-114 on HIV-1 Protease with Highly Drug Resistant Mutations D30N, I50V, and L90M.
J.Med.Chem., 49, 2006
|
|
2F93
 
 | | K Intermediate Structure of Sensory Rhodopsin II/Transducer Complex in Combination with the Ground State Structure | | Descriptor: | RETINAL, Sensory rhodopsin II, Sensory rhodopsin II transducer, ... | | Authors: | Moukhametzianov, R.I, Klare, J.P, Efremov, R.G, Baecken, C, Goeppner, A, Labahn, J, Engelhard, M, Bueldt, G, Gordeliy, V.I. | | Deposit date: | 2005-12-05 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of the signal in sensory rhodopsin and its transfer to the cognate transducer.
Nature, 440, 2006
|
|
8SFJ
 
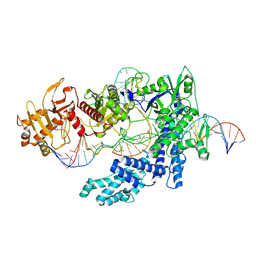 | | WT CRISPR-Cas12a with a 10bp R-loop | | Descriptor: | CRISPR-associated endonuclease Cas12a, DNA (5'-D(P*CP*AP*CP*TP*TP*AP*TP*CP*AP*CP*TP*AP*AP*AP*AP*GP*AP*TP*CP*GP*GP*AP*AP*G)-3'), DNA (5'-D(P*CP*TP*TP*CP*CP*GP*AP*TP*CP*TP*TP*TP*TP*AP*GP*TP*GP*AP*T)-3'), ... | | Authors: | Strohkendl, I, Taylor, D.W. | | Deposit date: | 2023-04-11 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | WT CRISPR-Cas12a with a 10bp R-loop
To Be Published
|
|
8SFH
 
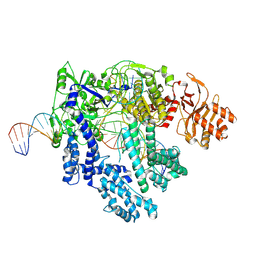 | |
1AI1
 
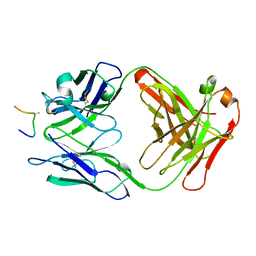 | | HIV-1 V3 LOOP MIMIC | | Descriptor: | AIB142, IGG1-KAPPA 59.1 FAB (HEAVY CHAIN), IGG1-KAPPA 59.1 FAB (LIGHT CHAIN) | | Authors: | Ghiara, J.B, Wilson, I.A. | | Deposit date: | 1996-11-06 | | Release date: | 1997-05-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of a constrained peptide mimic of the HIV-1 V3 loop neutralization site.
J.Mol.Biol., 266, 1997
|
|
8SFL
 
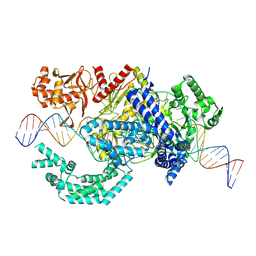 | |
1A6G
 
 | | CARBONMONOXY-MYOGLOBIN, ATOMIC RESOLUTION | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Vojtechovsky, J, Chu, K, Berendzen, J, Sweet, R.M, Schlichting, I. | | Deposit date: | 1998-02-25 | | Release date: | 1998-10-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structures of myoglobin-ligand complexes at near-atomic resolution.
Biophys.J., 77, 1999
|
|
8SFP
 
 | |
2RQG
 
 | | Structure of GSPT1/ERF3A-PABC | | Descriptor: | G1 to S phase transition 1, Polyadenylate-binding protein 1 | | Authors: | Osawa, M, Nakanishi, T, Hosoda, N, Uchida, S, Hoshino, T, Katada, I, Shimada, I. | | Deposit date: | 2009-05-08 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Eukaryotic Translation Termination Factor Gspt/Erf3 Recognizes Pabp with Chemical Exchange Using Two Overlapping Motifs
To be Published
|
|
