8OYP
 
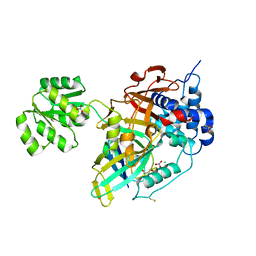 | | Crystal structure of Ubiquitin specific protease 11 (USP11) in complex with a substrate mimetic | | Descriptor: | CADMIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Maurer, S.K, Caulton, S.G, Ward, S.J, Emsley, J, Dreveny, I. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Ubiquitin-specific protease 11 structure in complex with an engineered substrate mimetic reveals a molecular feature for deubiquitination selectivity.
J.Biol.Chem., 299, 2023
|
|
2JJX
 
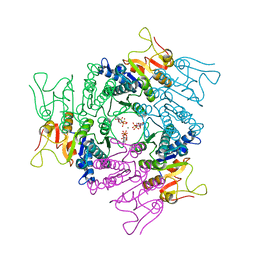 | | THE CRYSTAL STRUCTURE OF UMP KINASE FROM BACILLUS ANTHRACIS (BA1797) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, URIDYLATE KINASE | | Authors: | Meier, C, Carter, L.G, Mancini, E.J, Owens, R.J, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2008-04-23 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | The Crystal Structure of Ump Kinase from Bacillus Anthracis (Ba1797) Reveals an Allosteric Nucleotide-Binding Site.
J.Mol.Biol., 381, 2008
|
|
6WFW
 
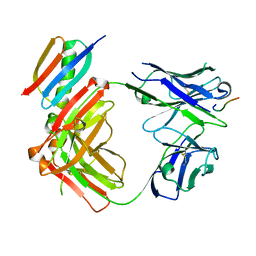 | | Crystal structure of Fab364 in complex with NPNA2 peptide from circumsporozoite protein | | Descriptor: | Fab364 heavy chain, Fab364 light chain, Immunoglobulin G-binding protein G, ... | | Authors: | Pholcharee, T, Oyen, D, Wilson, I.A. | | Deposit date: | 2020-04-04 | | Release date: | 2020-07-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Structural and biophysical correlation of anti-NANP antibodies with in vivo protection against P. falciparum.
Nat Commun, 12, 2021
|
|
2JLG
 
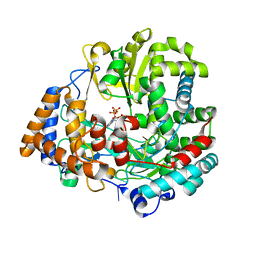 | | STRUCTURAL EXPLANATION FOR THE ROLE OF MN IN THE ACTIVITY OF PHI6 RNA-DEPENDENT RNA POLYMERASE | | Descriptor: | 5'-D(*DT DT DT DC DCP)-3', GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Poranen, M.M, Salgado, P.S, Koivunen, M.R.L, Wright, S, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2008-09-09 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Explanation for the Role of Mn2+ in the Activity of {Phi}6 RNA-Dependent RNA Polymerase.
Nucleic Acids Res., 36, 2008
|
|
4K9J
 
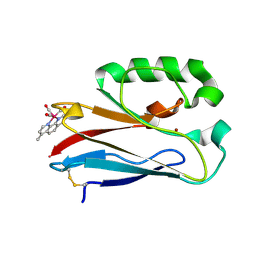 | | Structure of Re(CO)3(4,7-dimethyl-phen)(Thr126His)(Lys122Trp)(His83Glu)(Trp48Phe)(Tyr72Phe)(Tyr108Phe)AzCu(II), a Rhenium modified Azurin mutant | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COPPER (II) ION | | Authors: | Takematsu, K, Williamson, H.R, Blanco-Rodriguez, A.M, Sokolova, L, Nikolovski, P, Kaiser, J.T, Towrie, M, Clark, I.P, Vlcek Jr, A, Winkler, J.R, Gray, H.B. | | Deposit date: | 2013-04-20 | | Release date: | 2013-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tryptophan-accelerated electron flow across a protein-protein interface.
J.Am.Chem.Soc., 135, 2013
|
|
1FMM
 
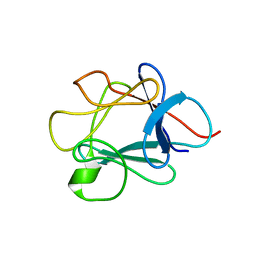 | | SOLUTION STRUCTURE OF NFGF-1 | | Descriptor: | ACIDIC FIBROBLAST GROWTH FACTOR | | Authors: | Arunkumar, A.I, Srisailam, S, Kumar, T.K.S, Chiu, I.M, Yu, C. | | Deposit date: | 2000-08-18 | | Release date: | 2001-08-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and stability of an acidic fibroblast growth factor from Notophthalmus viridescens.
J.Biol.Chem., 277, 2002
|
|
7SAV
 
 | | Native mu-conotoxin KIIIA isomer | | Descriptor: | Mu-conotoxin KIIIA | | Authors: | Schroeder, C.I, Tran, H.N.T. | | Deposit date: | 2021-09-23 | | Release date: | 2022-05-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and functional insights into the inhibition of human voltage-gated sodium channels by mu-conotoxin KIIIA disulfide isomers.
J.Biol.Chem., 298, 2022
|
|
8OSW
 
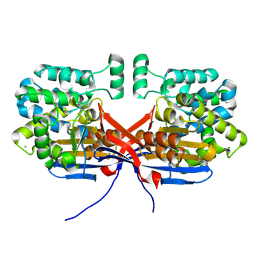 | | Crystal structure of Rhizobium etli L-asparaginase ReAIV (R4mC-1) | | Descriptor: | CHLORIDE ION, Putative L-asparaginase II protein, ZINC ION | | Authors: | Loch, J.I, Worsztynowicz, P, Sliwiak, J, Imiolczyk, B, Grzechowiak, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2023-04-20 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Rhizobium etli has two L-asparaginases with low sequence identity but similar structure and catalytic center.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8ONN
 
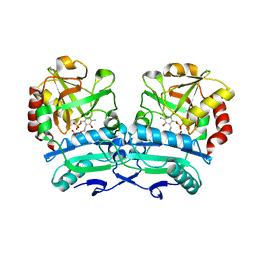 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant E113A complexed with 3-aminooxypropionic acid | | Descriptor: | 3-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]oxypropanoic acid, Aminotransferase class IV | | Authors: | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2023-04-03 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
1RJD
 
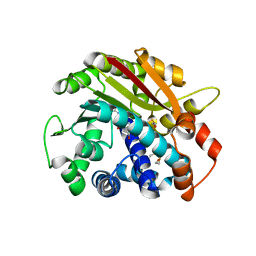 | | Structure of PPM1, a leucine carboxy methyltransferase involved in the regulation of protein phosphatase 2A activity | | Descriptor: | BETA-MERCAPTOETHANOL, S-ADENOSYLMETHIONINE, SULFATE ION, ... | | Authors: | Leulliot, N, Quevillon-Cheruel, S, Sorel, I, Li de La Sierra-Gallay, I, Collinet, B, Graille, M, Blondeau, K, Bettache, N, Poupon, A, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2003-11-19 | | Release date: | 2003-12-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of protein phosphatase methyltransferase 1 (PPM1), a leucine carboxyl methyltransferase involved in the regulation of protein phosphatase 2A activity
J.Biol.Chem., 279, 2004
|
|
8ORI
 
 | | Crystal structure of Rhizobium etli L-asparaginase ReAIV (orthorhombic) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Loch, J.I, Worsztynowicz, P, Sliwiak, J, Imiolczyk, B, Grzechowiak, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2023-04-14 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Rhizobium etli has two L-asparaginases with low sequence identity but similar structure and catalytic center.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5NWS
 
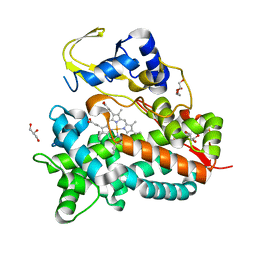 | | Crystal structure of saAcmM involved in actinomycin biosynthesis | | Descriptor: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, TETRAETHYLENE GLYCOL, ... | | Authors: | Driller, R, Semsary, S, Crnovicic, I, Vater, J, Keller, U, Loll, B. | | Deposit date: | 2017-05-08 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.227 Å) | | Cite: | Ketonization of Proline Residues in the Peptide Chains of Actinomycins by a 4-Oxoproline Synthase.
Chembiochem, 19, 2018
|
|
8P8J
 
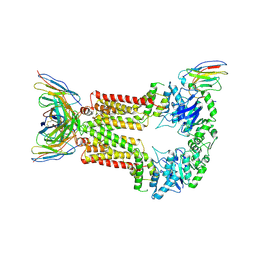 | | Structure of 5D3-Fab and nanobody(Nb96)-bound ABCG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3(Fab) heavy chain variable domain, 5D3(Fab) light chain variable domain, ... | | Authors: | Irobalieva, R.N, Manolaridis, I, Jackson, S.M, Ni, D, Pardon, E, Stahlberg, H, Steyaert, J, Locher, K.P. | | Deposit date: | 2023-06-01 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural Basis of the Allosteric Inhibition of Human ABCG2 by Nanobodies.
J.Mol.Biol., 435, 2023
|
|
2JAP
 
 | | Clavulanic Acid Dehydrogenase: Structural and Biochemical Analysis of the Final Step in the Biosynthesis of the beta-Lactamase Inhibitor Clavulanic acid | | Descriptor: | (2R,3Z,5R)-3-(2-HYDROXYETHYLIDENE)-7-OXO-4-OXA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, CLAVALDEHYDE DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | MacKenzie, A.K, Kershaw, N.J, Hernandez, H, Robinson, C.V, Schofield, C.J, Andersson, I. | | Deposit date: | 2006-11-29 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Clavulanic Acid Dehydrogenase: Structural and Biochemical Analysis of the Final Step in the Biosynthesis of the Beta-Lactamase Inhibitor Clavulanic Acid
Biochemistry, 46, 2007
|
|
5YP2
 
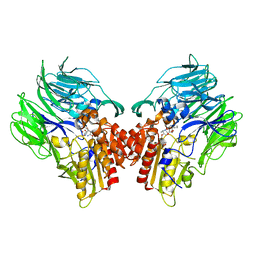 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) with DPP4 inhibitor from Pseudoxanthomonas mexicana WO24 | | Descriptor: | (2S,5R)-1-[2-[[1-(hydroxymethyl)cyclopentyl]amino]ethanoyl]pyrrolidine-2,5-dicarbonitrile, Dipeptidyl aminopeptidase 4, GLYCEROL | | Authors: | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
6QGU
 
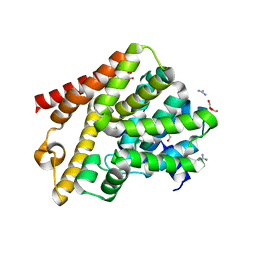 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-1361 | | Descriptor: | 5-(3-(cyclopentyloxy)-4-methoxyphenyl)-2-isopropyl-4,4-dimethyl-2,4-dihydro-3H-pyrazol-3-one, FORMIC ACID, GLYCEROL, ... | | Authors: | Singh, A.K, Blaazer, A.R, Zara, L, de Esch, I.J.P, Leurs, R, Brown, D.G. | | Deposit date: | 2019-01-13 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-1361
To be published
|
|
1D1H
 
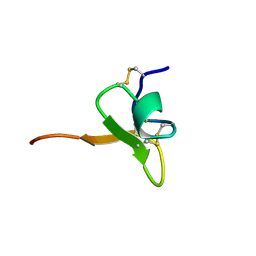 | | SOLUTION STRUCTURE OF HANATOXIN 1 | | Descriptor: | HANATOXIN TYPE 1 | | Authors: | Takahashi, H, Kim, J.I, Sato, K, Swartz, K.J, Shimada, I. | | Deposit date: | 1999-09-16 | | Release date: | 2000-09-20 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hanatoxin1, a gating modifier of voltage-dependent K(+) channels: common surface features of gating modifier toxins.
J.Mol.Biol., 297, 2000
|
|
2BXT
 
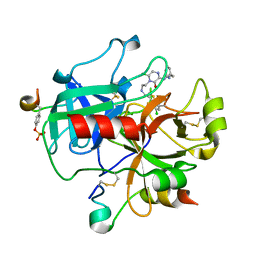 | | Design and Discovery of Novel, Potent Thrombin Inhibitors with a Solubilizing Cationic P1-P2-Linker | | Descriptor: | 6-CHLORO-1-(2-{[(5-CHLORO-1-BENZOTHIEN-3-YL)METHYL]AMINO}ETHYL)-3-[(2-PYRIDIN-2-YLETHYL)AMINO]-1,4-DIHYDROPYRAZIN-2-OL, ALPHA THROMBIN, HIRUDIN VARIANT-2 | | Authors: | Bulat, S, Bosio, S, Grabowski, E, Papadopoulos, M.A, Cerezo-Galvez, S, Rosenbaum, C, Matassa, V.G, Ott, I, Metz, G, Schamberger, J, Sekul, R, Feurer, A. | | Deposit date: | 2005-07-27 | | Release date: | 2006-10-26 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Design and Discovery of Novel, Potent Pyrazinone-Based Thrombin Inhibitors with a Solubilizing P1-P2-Linker
Lett.Drug Des.Discovery, 3, 2006
|
|
2JJV
 
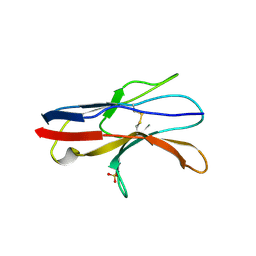 | | Structure of human signal regulatory protein (sirp) beta(2) | | Descriptor: | CHLORIDE ION, SIGNAL-REGULATORY PROTEIN BETA 1., SULFATE ION | | Authors: | Hatherley, D, Graham, S.C, Turner, J, Harlos, K, Stuart, D.I, Barclay, A.N. | | Deposit date: | 2008-04-22 | | Release date: | 2008-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Paired Receptor Specificity Explained by Structures of Signal Regulatory Proteins Alone and Complexed with Cd47.
Mol.Cell, 31, 2008
|
|
6J85
 
 | | Crystal structure of HinD apo | | Descriptor: | Nocardicin N-oxygenase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fei, H, Mori, T, Abe, I. | | Deposit date: | 2019-01-18 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis for the P450-catalyzed C-N bond formation in indolactam biosynthesis.
Nat.Chem.Biol., 15, 2019
|
|
8P7V
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | Descriptor: | 1,2-ETHANEDIOL, 1-ethyl-1-methyl-cyclohexane, FORMIC ACID, ... | | Authors: | Dym, O, Aggawal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | Deposit date: | 2023-05-31 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.737 Å) | | Cite: | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8P7I
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | Descriptor: | (2~{S},6~{R})-2,6-dimethyl-1,3-dioxane-4,4-diol, FORMIC ACID, Parathion hydrolase, ... | | Authors: | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | Deposit date: | 2023-05-30 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
2XV0
 
 | | Pseudomonas aeruginosa Azurin with mutated metal-binding loop sequence (CAAHAAM), chemically reduced, pH4.8 | | Descriptor: | AZURIN, COPPER (I) ION | | Authors: | Li, C, Sato, K, Monari, S, Salard, I, Sola, M, Banfield, M.J, Dennison, C. | | Deposit date: | 2010-10-22 | | Release date: | 2010-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Metal-Binding Loop Length is a Determinant of the Pka of a Histidine Ligand at a Type 1 Copper Site
Inorg.Chem., 50, 2011
|
|
8P7N
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | Descriptor: | FORMIC ACID, Parathion hydrolase, ZINC ION | | Authors: | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | Deposit date: | 2023-05-30 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
2JJD
 
 | | Protein Tyrosine Phosphatase, Receptor Type, E isoform | | Descriptor: | CHLORIDE ION, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE EPSILON | | Authors: | Elkins, J.M, Ugochukwu, E, Alfano, I, Barr, A.J, Bunkoczi, G, King, O.N.F, Filippakopoulos, P, Savitsky, P, Salah, E, Pike, A, Johansson, C, Das, S, Burgess-Brown, N.A, Gileadi, O, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Knapp, S. | | Deposit date: | 2008-03-31 | | Release date: | 2008-04-08 | | Last modified: | 2012-06-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Large-Scale Structural Analysis of the Classical Human Protein Tyrosine Phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
