1T4G
 
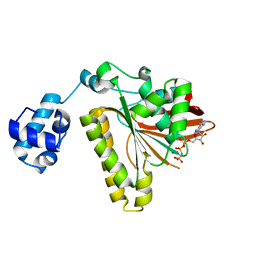 | | ATPase in complex with AMP-PNP | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wu, Y, He, Y, Moya, I.A, Qian, X, Luo, Y. | | Deposit date: | 2004-04-29 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Archaeal Recombinase RadA; A Snapshot of Its Extended Conformation.
Mol.Cell, 15, 2004
|
|
1TH1
 
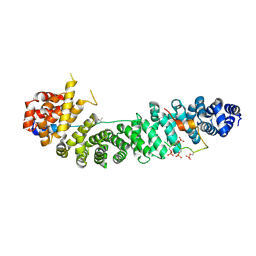 | | Beta-catenin in complex with a phosphorylated APC 20aa repeat fragment | | Descriptor: | Adenomatous polyposis coli protein, Beta-catenin | | Authors: | Xing, Y, Clements, W.K, Le Trong, I, Hinds, T.R, Stenkamp, R, Kimelman, D, Xu, W. | | Deposit date: | 2004-05-31 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a beta-Catenin/APC Complex Reveals a Critical Role for APC Phosphorylation in APC Function.
Mol.Cell, 15, 2004
|
|
2NZ2
 
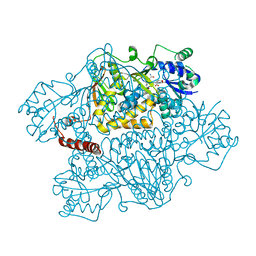 | | Crystal structure of human argininosuccinate synthase in complex with aspartate and citrulline | | Descriptor: | ASPARTIC ACID, Argininosuccinate synthase, CITRULLINE, ... | | Authors: | Karlberg, T, Uppenberg, J, Arrowsmith, C, Berglund, H, Busam, R.D, Collins, R, Edwards, A, Ericsson, U.B, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Hogbom, M, Johansson, I, Kotenyova, T, Magnusdottir, A, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Wallden, K, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-22 | | Release date: | 2006-12-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human argininosuccinate synthetase.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2YPJ
 
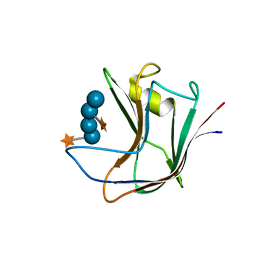 | | Non-catalytic carbohydrate binding module CBM65B | | Descriptor: | ENDOGLUCANASE CEL5A, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Luis, A.S, Venditto, I, Prates, J.A.M, Ferreira, L.M.A, Temple, M.J, Rogowski, A, Basle, A, Xue, J, Knox, J.P, Gilbert, H.J, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2012-10-30 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Understanding How Non-Catalytic Carbohydrate Binding Modules Can Display Specificity for Xyloglucan
J.Biol.Chem., 288, 2013
|
|
2Y80
 
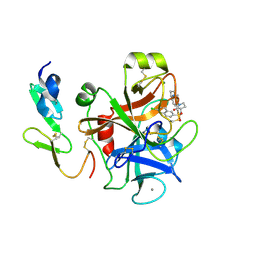 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with aminoindane and phenylpyrrolidine P4 motifs | | Descriptor: | 6-CHLORO-N-{(3S)-1-[(1S)-1-(DIMETHYLAMINO)-2,3-DIHYDRO-1H-INDEN-5-YL]-2-OXO-3-PYRROLIDINYL}-2-NAPHTHALENESULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Young, R.J, Adams, C, Blows, M, Brown, D, Burns-Kurtis, C.L, Chaudry, L, Chan, C, Convery, M.A, Davies, D.E, Exall, A.M, Foster, G, Harling, J.D, Hortense, E, Irving, W.R, Irvine, S, Jackson, S, Kleanthous, S, Pateman, A.J, Patikis, A.N, Roethka, T.J, Senger, S, Stelman, G.J, Toomey, J.R, West, R.I, Whittaker, C, Zhou, P, Watson, N.S. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-16 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Aminoindane and Phenylpyrrolidine P4 Motifs.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2D5K
 
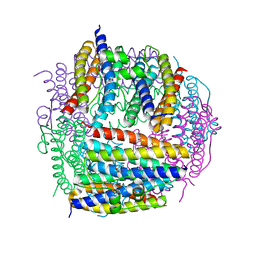 | | Crystal structure of Dps from Staphylococcus aureus | | Descriptor: | Dps family protein, GLYCEROL | | Authors: | Tanaka, Y, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2005-11-02 | | Release date: | 2006-10-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Nucleoid compaction by MrgA(Asp56Ala/Glu60Ala) does not contribute to staphylococcal cell survival against oxidative stress and phagocytic killing by macrophages
FEMS Microbiol. Lett., 360, 2014
|
|
2D47
 
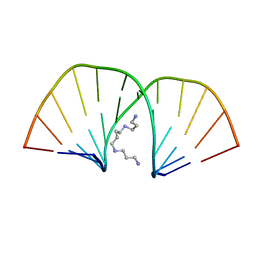 | | MOLECULAR STRUCTURE OF A COMPLETE TURN OF A-DNA | | Descriptor: | DNA (5'-D(*CP*CP*CP*CP*CP*GP*CP*GP*GP*GP*GP*G)-3'), SPERMINE | | Authors: | Verdaguer, N, Aymami, J, Fernandez-Forner, D, Fita, I, Coll, M, Huynh-Dinh, T, Igolen, J, Subirana, J.A. | | Deposit date: | 1991-10-02 | | Release date: | 1991-10-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular structure of a complete turn of A-DNA.
J.Mol.Biol., 221, 1991
|
|
2YSS
 
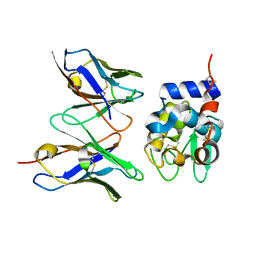 | | Crystal structure of Humanized HYHEL-10 FV mutant(HQ39KW47Y)-HEN lysozyme complex | | Descriptor: | ANTI-LYSOZYME ANTIBODY FV REGION, Lysozyme C | | Authors: | Nakanishi, T, Tsumoto, K, Yokota, A, Kondo, H, Kumagai, I. | | Deposit date: | 2007-04-03 | | Release date: | 2008-04-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Critical contribution of VH-VL interaction to reshaping of an antibody: the case of humanization of anti-lysozyme antibody, HyHEL-10
Protein Sci., 17, 2008
|
|
2YN2
 
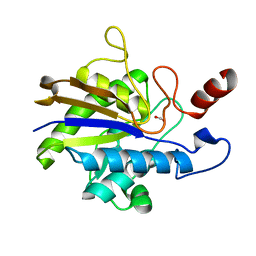 | | Huf protein - paralogue of the tau55 histidine phosphatase domain | | Descriptor: | FORMIC ACID, UNCHARACTERIZED PROTEIN YNL108C | | Authors: | Taylor, N.M.I, Glatt, S, Hennrich, M, von Scheven, G, Grotsch, H, Fernandez-Tornero, C, Rybin, V, Gavin, A.C, Kolb, P, Muller, C.W. | | Deposit date: | 2012-10-11 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and Functional Characterization of a Phosphatase Domain within Yeast General Transcription Factor Tfiiic.
J.Biol.Chem., 288, 2013
|
|
2DBE
 
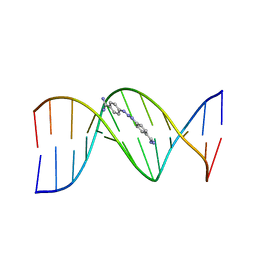 | | CRYSTAL STRUCTURE OF A BERENIL-DODECANUCLEOTIDE COMPLEX: THE ROLE OF WATER IN SEQUENCE-SPECIFIC LIGAND BINDING | | Descriptor: | BERENIL, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Brown, D.G, Sanderson, M.R, Skelly, J.V, Jenkins, T.C, Brown, T, Garman, E, Stuart, D.I, Neidle, S. | | Deposit date: | 1990-03-19 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a berenil-dodecanucleotide complex: the role of water in sequence-specific ligand binding.
EMBO J., 9, 1990
|
|
2YIJ
 
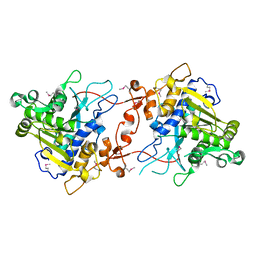 | | Crystal Structure of phospholipase A1 | | Descriptor: | PHOSPHOLIPASE A1-IIGAMMA | | Authors: | Lee, I. | | Deposit date: | 2011-05-13 | | Release date: | 2012-05-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Phospholipase A1
To be Published
|
|
2YN0
 
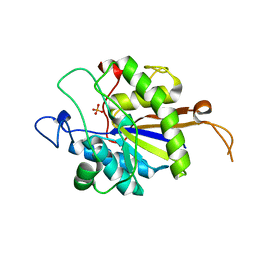 | | tau55 histidine phosphatase domain | | Descriptor: | PHOSPHATE ION, TRANSCRIPTION FACTOR TAU 55 KDA SUBUNIT | | Authors: | Taylor, N.M.I, Glatt, S, Hennrich, M, von Scheven, G, Grotsch, H, Fernandez-Tornero, C, Rybin, V, Gavin, A.C, Kolb, P, Muller, C.W. | | Deposit date: | 2012-10-11 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Characterization of a Phosphatase Domain within Yeast General Transcription Factor Iiic.
J.Biol.Chem., 288, 2013
|
|
2YIB
 
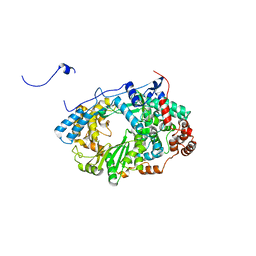 | | Structure of the RNA polymerase VP1 from Infectious Pancreatic Necrosis Virus | | Descriptor: | RNA-DIRECTED RNA POLYMERASE | | Authors: | Graham, S.C, Sarin, L.P, Bahar, M.W, Myers, R.A, Stuart, D.I, Bamford, D.H, Grimes, J.M. | | Deposit date: | 2011-05-11 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The N-Terminus of the RNA Polymerase from Infectious Pancreatic Necrosis Virus is the Determinant of Genome Attachment.
Plos Pathog., 7, 2011
|
|
2DKJ
 
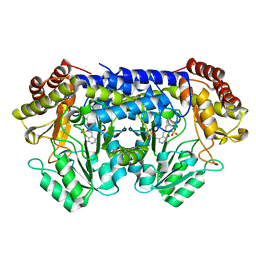 | | Crystal Structure of T.th.HB8 Serine Hydroxymethyltransferase | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SULFATE ION, serine hydroxymethyltransferase | | Authors: | Kai, K, Goto, M, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-11 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal Structure of T.th.HB8 Serine Hydroxymethyltransferase
To be Published
|
|
2YRR
 
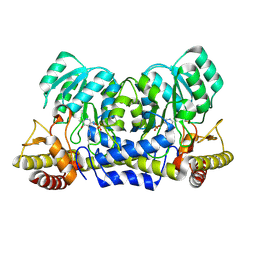 | | hypothetical alanine aminotransferase (TTH0173) from Thermus thermophilus HB8 | | Descriptor: | Aminotransferase, class V, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Miyahara, I, Matsumura, M, Goto, M, Omi, R, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-02 | | Release date: | 2008-04-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | hypothetical alanine aminotransferase (TTH0173) from Thermus thermophilus HB8
To be Published
|
|
2DF4
 
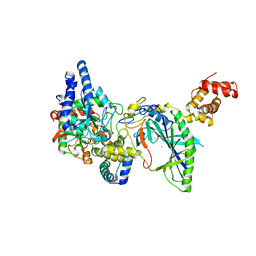 | | Structure of tRNA-Dependent Amidotransferase GatCAB complexed with Mn2+ | | Descriptor: | Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit C, Glutamyl-tRNA(Gln) amidotransferase subunit A, ... | | Authors: | Nakamura, A, Yao, M, Tanaka, I. | | Deposit date: | 2006-02-23 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Ammonia channel couples glutaminase with transamidase reactions in GatCAB
Science, 312, 2006
|
|
2Y6F
 
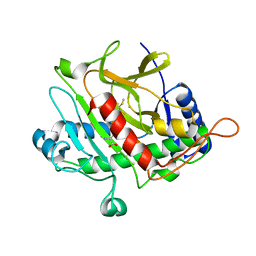 | | Isopenicillin N synthase with AC-D-S-methyl-3R-methylcysteine | | Descriptor: | (2S)-2-AMINO-6-[[(2R)-1-[[(2S)-1-HYDROXY-3-METHYLSULFANYL-1-OXO-BUTAN-2-YL]AMINO]-1-OXO-3-SULFANYL-PROPAN-2-YL]AMINO]-6-OXO-HEXANOIC ACID, FE (III) ION, ISOPENICILLIN N SYNTHASE, ... | | Authors: | Rutledge, P.J, Clifton, I.J, Ge, W. | | Deposit date: | 2011-01-21 | | Release date: | 2011-08-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Isopenicillin N Synthase Binds Delta-(L-Alpha-Aminoadipoyl)-L-Cysteinyl-D-Thia-Allo-Isoleucine Through Both Sulfur Atoms.
Chembiochem, 12, 2011
|
|
2YGB
 
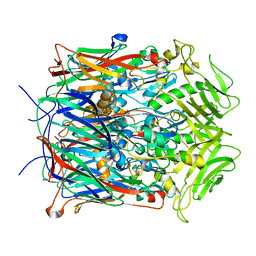 | |
2DG7
 
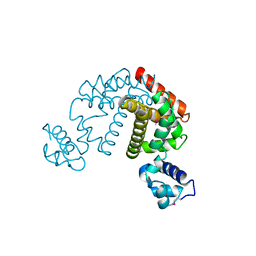 | | Crystal structure of the putative transcriptional regulator SCO0337 from Streptomyces coelicolor A3(2) | | Descriptor: | putative transcriptional regulator | | Authors: | Hayashi, T, Tanaka, Y, Sakai, N, Yao, M, Tamura, T, Tanaka, I. | | Deposit date: | 2006-03-08 | | Release date: | 2007-03-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the putative transcriptional regulator SCO0337 from Streptomyces coelicolor A3(2)
To be Published
|
|
2DQI
 
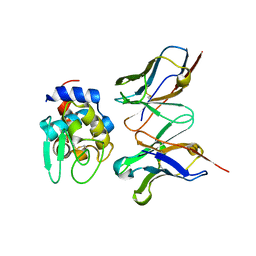 | | Crystal structure of hyhel-10 FV mutant (Ly50a) complexed with hen egg lysozyme | | Descriptor: | Ig VH,anti-lysozyme, Lysozyme C, lysozyme binding Ig kappa chain V23-J2 region | | Authors: | Shiroishi, M, Kondo, H, Tsumoto, K, Kumagai, I. | | Deposit date: | 2006-05-26 | | Release date: | 2007-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural consequences of mutations in interfacial Tyr residues of a protein antigen-antibody complex. The case of HyHEL-10-HEL
J.Biol.Chem., 282, 2007
|
|
2W6Q
 
 | | Crystal structure of Biotin carboxylase from E. coli in complex with the triazine-2,4-diamine fragment | | Descriptor: | 6-(2-phenoxyethoxy)-1,3,5-triazine-2,4-diamine, BIOTIN CARBOXYLASE, CHLORIDE ION | | Authors: | Mochalkin, I, Miller, J.R. | | Deposit date: | 2008-12-18 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Antibacterial Biotin Carboxylase Inhibitors by Virtual Screening and Fragment-Based Approaches.
Acs Chem.Biol., 4, 2009
|
|
2W71
 
 | |
2DVZ
 
 | | Structure of a periplasmic transporter | | Descriptor: | CADMIUM ION, GLUTAMIC ACID, Putative exported protein | | Authors: | Huvent, I, Belrhali, H, Antoine, R, Bompard, C, Locht, C, Jacob-Dubuisson, F, Villeret, V. | | Deposit date: | 2006-08-01 | | Release date: | 2006-11-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of Bordetella pertussis BugE solute receptor in a bound conformation
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2W96
 
 | | Crystal Structure of CDK4 in complex with a D-type cyclin | | Descriptor: | CELL DIVISION PROTEIN KINASE 4, G1/S-SPECIFIC CYCLIN-D1, GLYCEROL | | Authors: | Day, P.J, Cleasby, A, Tickle, I.J, Reilly, M.O, Coyle, J.E, Holding, F.P, McMenamin, R.L, Yon, J, Chopra, R, Lengauer, C, Jhoti, H. | | Deposit date: | 2009-01-21 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Human Cdk4 in Complex with a D-Type Cyclin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2WAM
 
 | | Crystal structure of Mycobacterium tuberculosis unknown function protein Rv2714 | | Descriptor: | CONSERVED HYPOTHETICAL ALANINE AND LEUCINE RICH PROTEIN, GLYCEROL | | Authors: | Bellinzoni, M, Grana, M, Buschiazzo, A, Miras, I, Haouz, A, Alzari, P.M. | | Deposit date: | 2009-02-09 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Mycobacterium Tuberculosis Rv2714, a Representative of a Duplicated Gene Family in Actinobacteria.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
