2BL8
 
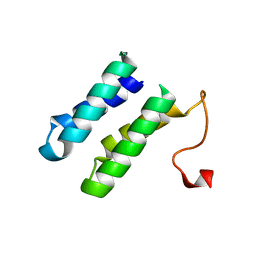 | | 1.6 Angstrom crystal structure of EntA-im: a bacterial immunity protein conferring immunity to the antimicrobial activity of the pediocin-like bacteriocin, enterocin A | | Descriptor: | CITRATE ANION, ENTEROCINE A IMMUNITY PROTEIN | | Authors: | Johnsen, L, Dalhus, B, Leiros, I, Nissen-Meyer, J. | | Deposit date: | 2005-03-02 | | Release date: | 2005-03-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.6-A Crystal Structure of Enta-Im: A Bacterial Immunity Protein Conferring Immunity to the Antimicrobial Activity of the Pediocin-Like Bacteriocin Enterocin A
J.Biol.Chem., 280, 2005
|
|
3OZ6
 
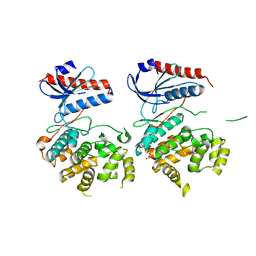 | | Crystal structure of MapK from Cryptosporidium Parvum, cgd2_1960 | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 1, serine/threonine protein kinase | | Authors: | Wernimont, A.K, Lew, J, Lin, Y.H, Sullivan, H, Hassanali, A, Kozieradzki, I, Cossar, D, Arrowsmith, C.H, Bochkarev, A, Bountra, C, Edwards, A.M, Weigelt, J, Hui, R, Neculai, A.M, Hutchinson, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-24 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of MapK from Cryptosporidium Parvum, cgd2_1960
To be Published
|
|
2BFW
 
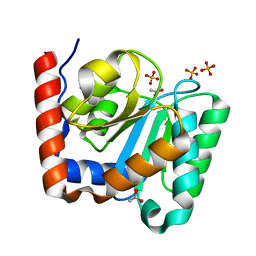 | | Structure of the C domain of glycogen synthase from Pyrococcus abyssi | | Descriptor: | ACETATE ION, GLGA GLYCOGEN SYNTHASE, SULFATE ION | | Authors: | Horcajada, C, Guinovart, J.J, Fita, I, Ferrer, J.C. | | Deposit date: | 2004-12-15 | | Release date: | 2005-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of an Archaeal Glycogen Synthase: Insights Into Oligomerisation and Substrate Binding of Eukaryotic Glycogen Synthases.
J.Biol.Chem., 281, 2006
|
|
1AEC
 
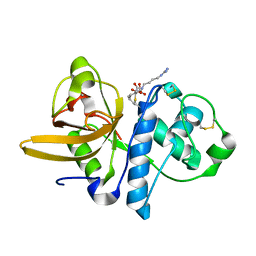 | | CRYSTAL STRUCTURE OF ACTINIDIN-E-64 COMPLEX+ | | Descriptor: | ACTINIDIN, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE | | Authors: | Varughese, K.I. | | Deposit date: | 1992-02-05 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of an actinidin-E-64 complex.
Biochemistry, 31, 1992
|
|
2BOO
 
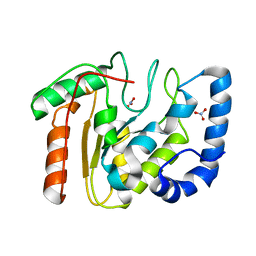 | | The crystal structure of Uracil-DNA N-Glycosylase (UNG) from Deinococcus radiodurans. | | Descriptor: | NITRATE ION, URACIL-DNA GLYCOSYLASE | | Authors: | Leiros, I, Moe, E, Smalas, A.O, McSweeney, S. | | Deposit date: | 2005-04-13 | | Release date: | 2005-07-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Uracil-DNA N-Glycosylase (Ung) from Deinococcus Radiodurans.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5D1Y
 
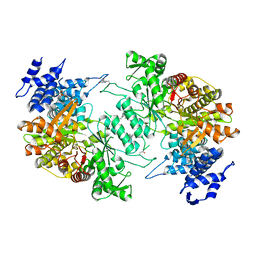 | | Low resolution crystal structure of human ribonucleotide reductase alpha6 hexamer in complex with dATP | | Descriptor: | Ribonucleoside-diphosphate reductase large subunit | | Authors: | Ando, N, Li, H, Brignole, E.J, Thompson, S, McLaughlin, M.I, Page, J, Asturias, F, Stubbe, J, Drennan, C.L. | | Deposit date: | 2015-08-04 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (9.005 Å) | | Cite: | Allosteric Inhibition of Human Ribonucleotide Reductase by dATP Entails the Stabilization of a Hexamer.
Biochemistry, 55, 2016
|
|
2KE6
 
 | |
2KJB
 
 | |
3OUD
 
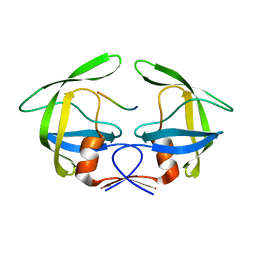 | | MDR769 HIV-1 protease complexed with CA/p2 hepta-peptide | | Descriptor: | CA/p2 substrate peptide, MDR HIV-1 protease | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
2KHU
 
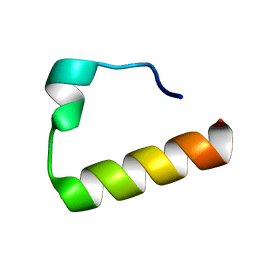 | | Solution Structure of the Ubiquitin-Binding Motif of Human Polymerase Iota | | Descriptor: | Immunoglobulin G-binding protein G, DNA polymerase iota | | Authors: | Bomar, M.G, D'Souza, S, Bienko, M, Dikic, I, Walker, G. | | Deposit date: | 2009-04-11 | | Release date: | 2010-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Unconventional Ubiquitin Recognition by the Ubiquitin-Binding Motif within the Y Family DNA Polymerases iota and Rev1.
Mol.Cell, 37, 2010
|
|
1B6K
 
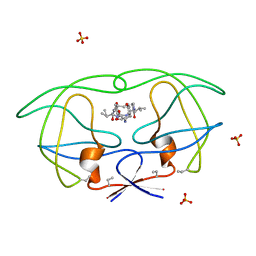 | | HIV-1 PROTEASE COMPLEXED WITH MACROCYCLIC PEPTIDOMIMETIC INHIBITOR 5 | | Descriptor: | N-[3-(8-SEC-BUTYL-7,10-DIOXO-2-OXA-6,9-DIAZA-BICYCLO[11.2.2]HEPTADECA-1(16),13(17),14- TRIEN-11-YLAMINO)-2-HYDROXY-1-(4-HYDROXY-BENZYL)-PROPYL]-3-METHYL-2- (2-OXO-PYRROLIDIN-1-YL)-BUTYRAMIDE, RETROPEPSIN, SULFATE ION | | Authors: | Martin, J.L, Begun, J, Schindeler, A, Wickramasinghe, W.A, Alewood, D, Alewood, P.F, Bergman, D.A, Brinkworth, R.I, Abbenante, G, March, D.R, Reid, R.C, Fairlie, D.P. | | Deposit date: | 1999-01-17 | | Release date: | 2000-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular recognition of macrocyclic peptidomimetic inhibitors by HIV-1 protease.
Biochemistry, 38, 1999
|
|
3P7I
 
 | |
2BSA
 
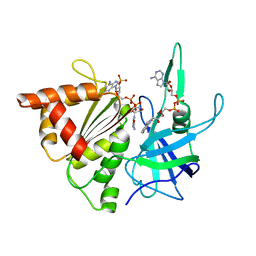 | | Ferredoxin-Nadp Reductase (Mutation: Y 303 S) complexed with NADP | | Descriptor: | FERREDOXIN-NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Maya, C.M, Hermoso, J.A, Perez-Dorado, I, Tejero, J, Julvez, M.M, Gomez-Moreno, C, Medina, M. | | Deposit date: | 2005-05-20 | | Release date: | 2005-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | C-Terminal Tyrosine of Ferredoxin-Nadp(+) Reductase in Hydride Transfer Processes with Nad(P)(+)/H.
Biochemistry, 44, 2005
|
|
2K6D
 
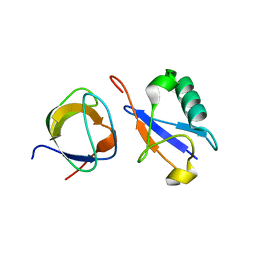 | | CIN85 Sh3-C domain in complex with ubiquitin | | Descriptor: | SH3 domain-containing kinase-binding protein 1, Ubiquitin | | Authors: | Forman-Kay, J, Bezsonova, I. | | Deposit date: | 2008-07-07 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Interactions between the three CIN85 SH3 domains and ubiquitin: implications for CIN85 ubiquitination.
Biochemistry, 47, 2008
|
|
2K8G
 
 | |
2K6L
 
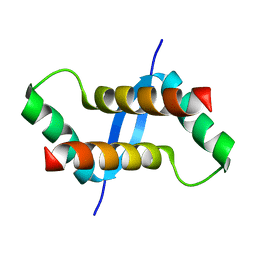 | | The solution structure of XACb0070 from Xanthonomas axonopodis pv citri reveals this new protein is a member of the RHH family of transcriptional repressors | | Descriptor: | Putative uncharacterized protein | | Authors: | Gallo, M, Cicero, D.O, Amata, I, Eliseo, T, Paci, M, Spisni, A, Ferrari, E, Pertinhez, T.A, Farah, C.S. | | Deposit date: | 2008-07-11 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure reveals that XACb0070 from the plant pathogen Xanthomonas citri belongs to the RHH superfamily of bacterial DNA-binding proteins
To be Published
|
|
2KEL
 
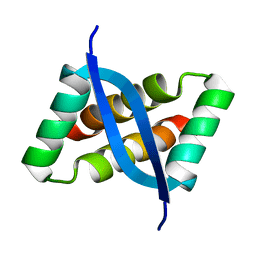 | | Structure of the transcription regulator SvtR from the hyperthermophilic archaeal virus SIRV1 | | Descriptor: | Uncharacterized protein 56B | | Authors: | Guilliere, F, Kessler, A, Peixeiro, N, Sezonov, G, Prangishvili, D, Delepierre, M, Guijarro, J.I. | | Deposit date: | 2009-01-30 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure, function, and targets of the transcriptional regulator SvtR from the hyperthermophilic archaeal virus SIRV1.
J.Biol.Chem., 284, 2009
|
|
2C2Q
 
 | | The crystal structure of mismatch specific uracil-DNA glycosylase (MUG) from Deinococcus radiodurans. Inactive mutant Asp93Ala. | | Descriptor: | ACETATE ION, G/U MISMATCH-SPECIFIC DNA GLYCOSYLASE | | Authors: | Moe, E, Leiros, I, Smalas, A.O, McSweeney, S. | | Deposit date: | 2005-09-29 | | Release date: | 2005-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Mismatch Specific Uracil-DNA Glycosylase (Mug) from Deinococcus Radiodurans Reveals a Novel Catalytic Residue and Broad Substrate Specificity
J.Biol.Chem., 281, 2006
|
|
2KC2
 
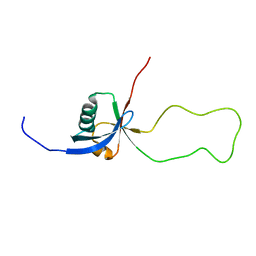 | | NMR structure of the F1 domain (residues 86-202) of the talin | | Descriptor: | Talin-1 | | Authors: | Goult, B.T, Elliott, P.R, Roberts, G.C.K, Critchely, D.R, Barsukov, I.L. | | Deposit date: | 2008-12-13 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of a double ubiquitin-like domain in the talin head: a role in integrin activation.
Embo J., 29, 2010
|
|
2BXE
 
 | | Human serum albumin complexed with diflunisal | | Descriptor: | 5-(2,4-DIFLUOROPHENYL)-2-HYDROXY-BENZOIC ACID, SERUM ALBUMIN | | Authors: | Ghuman, J, Zunszain, P.A, Petitpas, I, Bhattacharya, A.A, Curry, S. | | Deposit date: | 2005-07-26 | | Release date: | 2005-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Basis of the Drug-Binding Specificity of Human Serum Albumin.
J.Mol.Biol., 353, 2005
|
|
3MNW
 
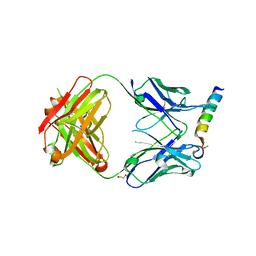 | | Crystal structure of the non-neutralizing HIV antibody 13H11 Fab fragment with a gp41 MPER-derived peptide in a helical conformation | | Descriptor: | 1,2-ETHANEDIOL, ANTI-HIV-1 ANTIBODY 13H11 HEAVY CHAIN, ANTI-HIV-1 ANTIBODY 13H11 LIGHT CHAIN, ... | | Authors: | Nicely, N.I, Dennison, S.M, Kelsoe, G, Liao, H.-X, Alam, S.M, Haynes, B.F. | | Deposit date: | 2010-04-22 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Non-Neutralizing HIV-1 gp41 Envelope Antibody Demonstrates Neutralization Mechanism of gp41 Antibodies
To be Published
|
|
3MOB
 
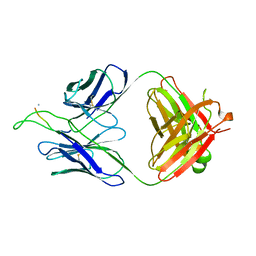 | | Crystal structure of the neutralizing HIV antibody 2F5 Fab fragment (recombinantly produced Fab) with 11 aa gp41 MPER-derived peptide | | Descriptor: | ANTI-HIV-1 ANTIBODY 2F5 HEAVY CHAIN, ANTI-HIV-1 ANTIBODY 2F5 LIGHT CHAIN, gp41 MPER-derived peptide | | Authors: | Nicely, N.I, Dennison, S.M, Kelsoe, G, Liao, H.-X, Alam, S.M, Haynes, B.F. | | Deposit date: | 2010-04-22 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of a Non-Neutralizing HIV-1 gp41 Envelope Antibody Demonstrates Neutralization Mechanism of gp41 Antibodies
To be Published
|
|
2KSE
 
 | | Backbone structure of the membrane domain of E. coli histidine kinase receptor QseC, Center for Structures of Membrane Proteins (CSMP) target 4311C | | Descriptor: | Sensor protein qseC | | Authors: | Maslennikov, I, Klammt, C, Kefala, G, Esquivies, L, Kwiatkowski, W, Choe, S, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2010-01-02 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Membrane domain structures of three classes of histidine kinase receptors by cell-free expression and rapid NMR analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MQM
 
 | | Crystal Structure of the Bromodomain of human ASH1L | | Descriptor: | Probable histone-lysine N-methyltransferase ASH1L | | Authors: | Filippakopoulos, P, Picaud, S, Keates, T, Felletar, I, Vollmar, M, Chaikuad, A, Krojer, T, Canning, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-28 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
2H3D
 
 | | Crystal Structure of Mouse Nicotinamide Phosphoribosyltransferase/Visfatin/Pre-B Cell Colony Enhancing Factor in Complex with Nicotinamide Mononuleotide | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Nicotinamide phosphoribosyltransferase | | Authors: | Wang, T, Zhang, X, Bheda, P, Revollo, J.R, Imai, S.I, Wolberger, C. | | Deposit date: | 2006-05-22 | | Release date: | 2006-06-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Nampt/PBEF/visfatin, a mammalian NAD(+) biosynthetic enzyme.
Nat.Struct.Mol.Biol., 13, 2006
|
|
