4K3X
 
 | |
1XMW
 
 | | CD3 EPSILON AND DELTA ECTODOMAIN FRAGMENT COMPLEX IN SINGLE-CHAIN CONSTRUCT | | Descriptor: | Chimeric CD3 mouse Epsilon and sheep Delta Ectodomain Fragment Complex | | Authors: | Sun, Z.-Y.J, Kim, S.T, Kim, I.C, Fahmy, A, Reinherz, E.L, Wagner, G. | | Deposit date: | 2004-10-04 | | Release date: | 2004-11-30 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CD3epsilondelta ectodomain and comparison with CD3epsilongamma as a basis for modeling T cell receptor topology and signaling.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
4HSH
 
 | | tRNA-guanine transglycosylase Y106F, V233G mutant in complex with queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Biela, I, Tidten-Luksch, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2012-10-30 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor.
Plos One, 8, 2013
|
|
3FO2
 
 | |
1V6V
 
 | | Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 3(2)-alpha-L-arabinofuranosyl-xylotriose | | Descriptor: | ENDO-1,4-BETA-D-XYLANASE, alpha-L-arabinofuranose-(1-3)-[beta-D-xylopyranose-(1-4)]beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, beta-D-xylopyranose, ... | | Authors: | Fujimoto, Z, Kaneko, S, Kuno, A, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2003-12-04 | | Release date: | 2004-04-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of decorated xylooligosaccharides bound to a family 10 xylanase from Streptomyces olivaceoviridis E-86
J.Biol.Chem., 279, 2004
|
|
3FO1
 
 | |
3FA2
 
 | |
3IO4
 
 | | Huntingtin amino-terminal region with 17 Gln residues - Crystal C90 | | Descriptor: | CALCIUM ION, Maltose-binding periplasmic protein,Huntingtin fusion protein, ZINC ION | | Authors: | Kim, M.W, Chelliah, Y, Kim, S.W, Otwinowski, Z, Bezprozvanny, I. | | Deposit date: | 2009-08-13 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.63 Å) | | Cite: | Secondary structure of Huntingtin amino-terminal region.
Structure, 17, 2009
|
|
1UZP
 
 | | Integrin binding cbEGF22-TB4-cbEGF33 fragment of human fibrillin-1, Sm bound form cbEGF23 domain only. | | Descriptor: | FIBRILLIN-1, SAMARIUM (III) ION | | Authors: | Lee, S.S.J, Knott, V, Harlos, K, Handford, P.A, Stuart, D.I. | | Deposit date: | 2004-03-15 | | Release date: | 2004-04-08 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of the Integrin Binding Fragment from Fibrillin-1 Gives New Insights Into Microfibril Organization
Structure, 12, 2004
|
|
4LYC
 
 | | Cd ions within a lysoyzme single crystal | | Descriptor: | CADMIUM ION, Lysozyme C | | Authors: | Wei, H, House, S, Wu, J, Zhang, J, Wang, Z, He, Y, Gao, Y.-G, Robinson, H, Li, W, Zuo, J.-M, Robertson, I.M, Lu, Y. | | Deposit date: | 2013-07-30 | | Release date: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Enhanced and tunable fluorescent quantum dots within a single crystal of protein
TO BE PUBLISHED
|
|
1ZKZ
 
 | | Crystal Structure of BMP9 | | Descriptor: | Growth/differentiation factor 2 | | Authors: | Brown, M.A, Zhao, Q, Baker, K.A, Naik, C, Chen, C, Pukac, L, Singh, M, Tsareva, T, Parice, Y, Mahoney, A, Roschke, V, Sanyal, I, Choe, S. | | Deposit date: | 2005-05-04 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of BMP-9 and functional interactions with pro-region and receptors
J.Biol.Chem., 280, 2005
|
|
3IFE
 
 | | 1.55 Angstrom Resolution Crystal Structure of Peptidase T (pepT-1) from Bacillus anthracis str. 'Ames Ancestor'. | | Descriptor: | Peptidase T, SODIUM ION, SULFATE ION, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-24 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.55 Angstrom Resolution Crystal Structure of Peptidase T (pepT-1) from Bacillus anthracis str. 'Ames Ancestor'.
To be Published
|
|
3PNQ
 
 | | Crystal Structure of E.coli Dha kinase DhaK (H56N) complex with Dha | | Descriptor: | Dihydroxyacetone, PTS-dependent dihydroxyacetone kinase, dihydroxyacetone-binding subunit dhaK | | Authors: | Shi, R, McDonald, L, Matte, A, Cygler, M, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-11-19 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and mechanistic insight into covalent substrate binding by Escherichia coli dihydroxyacetone kinase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1TNH
 
 | |
4I0P
 
 | | HLA-DO in complex with HLA-DM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | Guce, A.I, Mortimer, S.E, Stern, L.J. | | Deposit date: | 2012-11-18 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | HLA-DO acts as a substrate mimic to inhibit HLA-DM by a competitive mechanism.
Nat.Struct.Mol.Biol., 20, 2013
|
|
1TNJ
 
 | |
4HXF
 
 | | Acylaminoacyl peptidase in complex with Z-Gly-Gly-Phe-chloromethyl ketone | | Descriptor: | CHLORIDE ION, HEXANE-1,6-DIOL, MAGNESIUM ION, ... | | Authors: | Kiss-Szeman, A, Menyhard, D.K, Tichy-Racs, E, Hornung, B, Radi, K, Szeltner, Z, Domokos, K, Szamosi, I, Naray-Szabo, G, Polgar, L, Harmat, V. | | Deposit date: | 2012-11-09 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | A Self-compartmentalizing Hexamer Serine Protease from Pyrococcus Horikoshii: SUBSTRATE SELECTION ACHIEVED THROUGH MULTIMERIZATION.
J.Biol.Chem., 288, 2013
|
|
4HQS
 
 | | Crystal structure of the pneumoccocal exposed lipoprotein thioredoxin sp_0659 (Etrx1) from Streptococcus pneumoniae strain TIGR4 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Thioredoxin family protein | | Authors: | Bartual, S.G, Saleh, M, Abdullah, M.R, Jensch, I, Asmat, T.M, Petruschka, L, Pribyl, T, Hermoso, J.A, Hammerschmidt, S. | | Deposit date: | 2012-10-26 | | Release date: | 2013-12-25 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Molecular architecture of Streptococcus pneumoniae surface thioredoxin-fold lipoproteins crucial for extracellular oxidative stress resistance and maintenance of virulence.
EMBO Mol Med, 5, 2013
|
|
3IB3
 
 | | Crystal Structure of SACOL2612 - CocE/NonD family hydrolase from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, CocE/NonD family hydrolase, NICKEL (II) ION, ... | | Authors: | Domagalski, M.J, Chruszcz, M, Osinski, T, Skarina, T, Onopriyenko, O, Cymborowski, M, Shumilin, I.A, Savchenko, A, Edwards, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-15 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of SACOL2612 - CocE/NonD family hydrolase from Staphylococcus aureus
To be Published
|
|
1ZLR
 
 | | Factor XI catalytic domain complexed with 2-guanidino-1-(4-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)phenyl)ethyl nicotinate | | Descriptor: | (1R)-2-{[AMINO(IMINO)METHYL]AMINO}-1-{4-[(4R)-4-(HYDROXYMETHYL)-1,3,2-DIOXABOROLAN-2-YL]PHENYL}ETHYL NICOTINATE, Coagulation factor XI, GLYCEROL, ... | | Authors: | Lazarova, T.I, Jin, L, Rynkiewicz, M, Gorga, J.C, Bibbins, F, Meyers, H.V, Babine, R, Strickler, J. | | Deposit date: | 2005-05-09 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Synthesis and in vitro biological evaluation of aryl boronic acids as potential inhibitors of factor XIa.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2ZOQ
 
 | | Structural dissection of human mitogen-activated kinase ERK1 | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, Mitogen-activated protein kinase 3, SODIUM ION, ... | | Authors: | Kinoshita, T, Tada, T, Nakae, S, Yoshida, I. | | Deposit date: | 2008-06-01 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of human mono-phosphorylated ERK1 at Tyr204
Biochem.Biophys.Res.Commun., 377, 2008
|
|
3J16
 
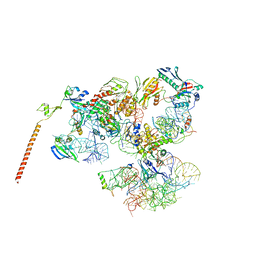 | | Models of ribosome-bound Dom34p and Rli1p and their ribosomal binding partners | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S24-A, ... | | Authors: | Becker, T, Franckenberg, S, Wickles, S, Shoemaker, C.J, Anger, A.M, Armache, J.-P, Sieber, H, Ungewickell, C, Berninghausen, O, Daberkow, I, Karcher, A, Thomm, M, Hopfner, K.-P, Green, R, Beckmann, R. | | Deposit date: | 2011-12-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Structural basis of highly conserved ribosome recycling in eukaryotes and archaea.
Nature, 482, 2012
|
|
1TZQ
 
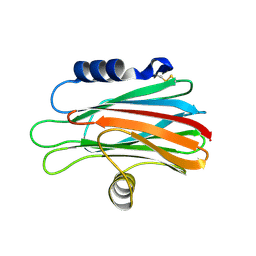 | | Crystal structure of the equinatoxin II 8-69 double cysteine mutant | | Descriptor: | Equinatoxin II | | Authors: | Kristan, K, Podlesek, Z, Hojnik, V, Gutirrez-Aguirre, I, Guncar, G, Turk, D.A, Gonzalez-Maas, J.M, Lakey, J.H, Anderluh, G. | | Deposit date: | 2004-07-11 | | Release date: | 2004-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Pore formation by equinatoxin, a eukaryotic pore-forming toxin, requires a flexible N-terminal region and a stable beta-sandwich
J.Biol.Chem., 279, 2004
|
|
4MHX
 
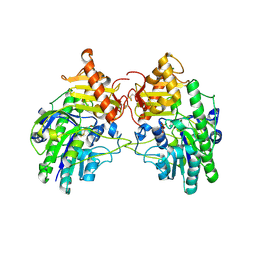 | | Crystal Structure of Sulfamidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Sidhu, N.S, Uson, I, Schreiber, K, Proepper, K, Becker, S, Gaertner, J, Kraetzner, R, Steinfeld, R, Sheldrick, G.M. | | Deposit date: | 2013-08-30 | | Release date: | 2014-05-14 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of sulfamidase provides insight into the molecular pathology of mucopolysaccharidosis IIIA.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1TNG
 
 | |
