4FWD
 
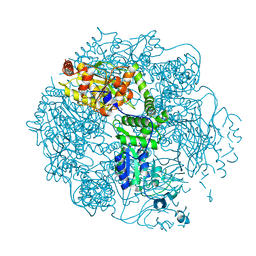 | | Crystal structure of the Lon-like protease MtaLonC in complex with bortezomib | | Descriptor: | N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, PHOSPHATE ION, TTC1975 peptidase | | Authors: | Chang, C.I, Kuo, C.I, Huang, K.F. | | Deposit date: | 2012-06-30 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of an ATP-independent Lon-like protease and its complexes with covalent inhibitors
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5NV6
 
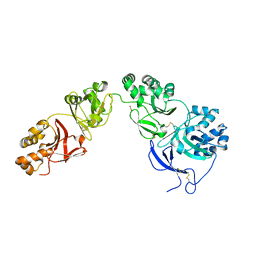 | | Structure of human transforming growth factor beta-induced protein (TGFBIp). | | Descriptor: | ACETATE ION, Transforming growth factor-beta-induced protein ig-h3 | | Authors: | Garcia-Castellanos, R, Nielsen, S.N, Runager, K, Thogersen, B.I, Goulas, T, Enghild, J.J, Gomis-Ruth, F.X. | | Deposit date: | 2017-05-03 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structural and Functional Implications of Human Transforming Growth Factor beta-Induced Protein, TGFBIp, in Corneal Dystrophies.
Structure, 25, 2017
|
|
2STB
 
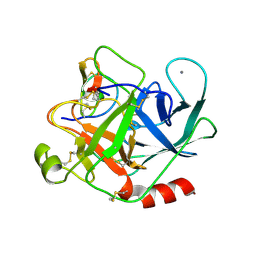 | | ANIONIC SALMON TRYPSIN IN COMPLEX WITH SQUASH SEED INHIBITOR (CUCURBITA PEPO TRYPSIN INHIBITOR II) | | Descriptor: | CALCIUM ION, PROTEIN (TRYPSIN INHIBITOR), PROTEIN (TRYPSIN) | | Authors: | Helland, R, Berglund, G.I, Otlewski, J, Apostoluk, W, Andersen, O.A, Willassen, N.P, Smalas, A.O. | | Deposit date: | 1998-12-11 | | Release date: | 2000-01-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of three new trypsin-squash-inhibitor complexes: a detailed comparison with other trypsins and their complexes.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
6DJ5
 
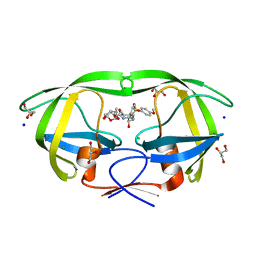 | | HIV-1 protease with mutation L76V in complex with GRL-0519 (tris-tetrahydrofuran as P2 ligand) | | Descriptor: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wong-Sam, A.E, Wang, Y.F, Weber, I.T. | | Deposit date: | 2018-05-24 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Drug Resistance Mutation L76V Alters Nonpolar Interactions at the Flap-Core Interface of HIV-1 Protease.
ACS Omega, 3, 2018
|
|
1K07
 
 | | Native FEZ-1 metallo-beta-lactamase from Legionella gormanii | | Descriptor: | ACETATE ION, FEZ-1 beta-lactamase, GLYCEROL, ... | | Authors: | Garcia-Saez, I, Mercuri, P.S, Kahn, R, Papamicael, C, Frere, J.M, Galleni, M, Dideberg, O. | | Deposit date: | 2001-09-18 | | Release date: | 2003-01-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Three-dimensional Structure of FEZ-1, a Monomeric Subclass B3 Metallo-[beta]-lactamase from Fluoribacter gormanii, in Native Form and in Complex with -Captopril
J.MOL.BIOL., 325, 2003
|
|
1GVI
 
 | | Thermus maltogenic amylase in complex with beta-CD | | Descriptor: | Cycloheptakis-(1-4)-(alpha-D-glucopyranose), MALTOGENIC AMYLASE | | Authors: | Kim, M.-S, Kim, J.-I, Oh, B.-H. | | Deposit date: | 2002-02-14 | | Release date: | 2002-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Cyclomaltodextrinase, Neopullulanase, and Maltogenic Amylase are Nearly Indistinguishable from Each Other
J.Biol.Chem., 277, 2002
|
|
1LTX
 
 | | Structure of Rab Escort Protein-1 in complex with Rab geranylgeranyl transferase and isoprenoid | | Descriptor: | AAAA, CHLORIDE ION, FARNESYL, ... | | Authors: | Pylypenko, O, Rak, A, Reents, R, Niculae, A, Thoma, N.H, Waldmann, H, Schlichting, I, Goody, R.S, Alexandrov, K. | | Deposit date: | 2002-05-21 | | Release date: | 2003-05-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Rab Escort Protein-1 in Complex with Rab Geranylgeranyltransferase
Mol.Cell, 11, 2003
|
|
6DVY
 
 | |
4FW9
 
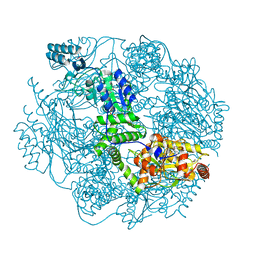 | | Crystal structure of the Lon-like protease MtaLonC | | Descriptor: | PHOSPHATE ION, TTC1975 peptidase | | Authors: | Chang, C.I, Ihara, K, Kuo, C.I, Huang, K.F, Wakatsuki, S. | | Deposit date: | 2012-06-30 | | Release date: | 2013-06-26 | | Last modified: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of an ATP-independent Lon-like protease and its complexes with covalent inhibitors
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3LDH
 
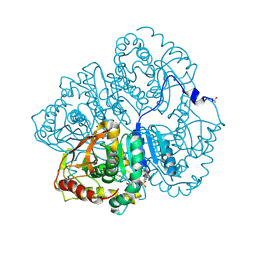 | | A comparison of the structures of apo dogfish m4 lactate dehydrogenase and its ternary complexes | | Descriptor: | LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PYRUVIC ACID | | Authors: | White, J.L, Hackert, M.L, Buehner, M, Adams, M.J, Ford, G.C, Lentzjunior, P.J, Smiley, I.E, Steindel, S.J, Rossmann, M.G. | | Deposit date: | 1974-06-06 | | Release date: | 1977-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A comparison of the structures of apo dogfish M4 lactate dehydrogenase and its ternary complexes.
J.Mol.Biol., 102, 1976
|
|
1DBJ
 
 | |
1KZ8
 
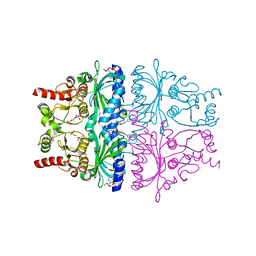 | | CRYSTAL STRUCTURE OF PORCINE FRUCTOSE-1,6-BISPHOSPHATASE COMPLEXED WITH A NOVEL ALLOSTERIC-SITE INHIBITOR | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE-1,6-BISPHOSPHATASE, ... | | Authors: | Wright, S.W, Carlo, A.A, Carty, M.D, Danley, D.E, Hageman, D.L, Karam, G.A, Levy, C.B, Mansour, M.N, Mathiowetz, A.M, McClure, L.D, Nestor, N.B, McPherson, R.K, Pandit, J, Pustilnik, L.R, Schulte, G.K, Soeller, W.C, Treadway, J.L, Wang, I.-K, Bauer, P.H. | | Deposit date: | 2002-02-06 | | Release date: | 2002-10-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ANILINOQUINAZOLINE INHIBITORS OF FRUCTOSE 1,6-BISPHOSPHATASE BIND AT A NOVEL ALLOSTERIC SITE: SYNTHESIS, IN VITRO CHARACTERIZATION, AND X-RAY CRYSTALLOGRAPHY
J.MED.CHEM., 45, 2002
|
|
5VTU
 
 | |
8FU3
 
 | | Structure Of Respiratory Syncytial Virus Polymerase with Novel Non-Nucleoside Inhibitor | | Descriptor: | 8-methoxy-3-methyl-N-{(2S)-3,3,3-trifluoro-2-[5-fluoro-6-(4-fluorophenyl)-4-(2-hydroxypropan-2-yl)pyridin-2-yl]-2-hydroxypropyl}cinnoline-6-carboxamide, Phosphoprotein, RNA-directed RNA polymerase L | | Authors: | Yu, X, Abeywickrema, P, Bonneux, B, Behera, I, Jacoby, E, Fung, A, Adhikary, S, Bhaumik, A, Carbajo, R.J, Bruyn, S.D, Miller, R, Patrick, A, Pham, Q, Piassek, M, Verheyen, N, Shareef, A, Sutto-Ortiz, P, Ysebaert, N, Vlijmen, H.V, Jonckers, T.H.M, Herschke, F, McLellan, J.S, Decroly, E, Fearns, R, Grosse, S, Roymans, D, Sharma, S, Rigaux, P, Jin, Z. | | Deposit date: | 2023-01-16 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural and mechanistic insights into the inhibition of respiratory syncytial virus polymerase by a non-nucleoside inhibitor.
Commun Biol, 6, 2023
|
|
6DLZ
 
 | | Open state GluA2 in complex with STZ after micelle signal subtraction | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Vassilevski, A.A, Sobolevsky, A.I. | | Deposit date: | 2018-06-04 | | Release date: | 2018-08-22 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanisms of Channel Block in Calcium-Permeable AMPA Receptors.
Neuron, 99, 2018
|
|
5TRQ
 
 | |
1H2Q
 
 | | Human CD55 domains 3 & 4 | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR | | Authors: | Williams, P, Chaudhry, Y, Goodfellow, I.G, Billington, J, Powell, R, Spiller, O.B, Evans, D.J, Lea, S.M. | | Deposit date: | 2002-08-13 | | Release date: | 2003-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
1H25
 
 | | CDK2/Cyclin A in complex with an 11-residue recruitment peptide from retinoblastoma-associated protein | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, RETINOBLASTOMA-ASSOCIATED PROTEIN | | Authors: | Tews, I, Cheng, K.Y, Lowe, E.D, Noble, M.E.M, Brown, N.R, Gul, S, Gamblin, S, Johnson, L.N. | | Deposit date: | 2002-07-31 | | Release date: | 2003-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Specificity Determinants of Recruitment Peptides Bound to Phospho-Cdk2/Cyclin A
Biochemistry, 41, 2002
|
|
1M38
 
 | | Structure of Inorganic Pyrophosphatase | | Descriptor: | COBALT (II) ION, INORGANIC PYROPHOSPHATASE, PHOSPHATE ION | | Authors: | Kuranova, I.P, Polyakov, K.M, Levdikov, V.M, Smirnova, E.A, Hohne, W.E, Lamzin, V.S, Meijers, R. | | Deposit date: | 2002-06-27 | | Release date: | 2003-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of Saccharomyces cerevisiae inorganic pyrophosphatase complexed with cobalt and phosphate ions
CRYSTALLOGRAPHY REPORTS, 48, 2003
|
|
8CXC
 
 | | Novel Anti-Mesothelin Antibodies Enable Crystallography of the Intact Mesothelin Ectodo- main and Engineering of Potent, T cell-engaging Bispecific Therapeutics | | Descriptor: | 3F2 Antibody heavy chain, 3F2 Antibody light chain, Mesothelin, ... | | Authors: | Bandaranayake, A.D, Rupert, P.B, Lin, I, Pilat, K, Ruff, R.O, Friend, D.J, Chan, M.K, Clarke, M, Carter, J, Meshinchi, S, Mehlin, C, Olson, J.M, Strong, R.K, Correnti, C.E. | | Deposit date: | 2022-05-20 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4.31 Å) | | Cite: | Novel mesothelin antibodies enable crystallography of the intact mesothelin ectodomain and engineering of potent, T cell-engaging bispecific therapeutics
Front. Drug Discov., 3, 2023
|
|
1H8H
 
 | | Bovine mitochondrial F1-ATPase crystallised in the presence of 5mm AMPPNP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, BOVINE MITOCHONDRIAL F1-ATPASE, ... | | Authors: | Braig, K, Menz, R.I, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2001-02-06 | | Release date: | 2001-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure and Nucleotide Occupancy of Bovine Mitochondrial F(1)-ATPase are not Influenced by Crystallisation at High Concentrations of Nucleotide
FEBS Lett., 494, 2001
|
|
1H2T
 
 | | Structure of the human nuclear cap-binding-complex (CBC) in complex with a cap analogue m7GpppG | | Descriptor: | 20 KDA NUCLEAR CAP BINDING PROTEIN, 7N-METHYL-8-HYDROGUANOSINE-5'-MONOPHOSPHATE, 80 KDA NUCLEAR CAP BINDING PROTEIN, ... | | Authors: | Mazza, C, Segref, A, Mattaj, I.W, Cusack, S. | | Deposit date: | 2002-08-16 | | Release date: | 2002-10-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Large-Scale Induced Fit Recognition of an M(7)Gpppg CAP Analogue by the Human Nuclear CAP-Binding Complex
Embo J., 21, 2002
|
|
4ZIU
 
 | | Crystal structure of native alpha-2-macroglobulin from Escherichia coli spanning the residues from domain MG7 to the C-terminus. | | Descriptor: | GLYCEROL, NICKEL (II) ION, Uncharacterized lipoprotein YfhM | | Authors: | Garcia-Ferrer, I, Arede, P, Gomez-Blanco, J, Luque, D, Duquerroy, S, Caston, J.R, Goulas, T, Gomis-Ruth, X.F. | | Deposit date: | 2015-04-28 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional insights into Escherichia coli alpha 2-macroglobulin endopeptidase snap-trap inhibition.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3LTN
 
 | | Inhibitor-stabilized topoisomerase IV-DNA cleavage complex (S. pneumoniae) | | Descriptor: | 3-amino-7-{(3R)-3-[(1S)-1-aminoethyl]pyrrolidin-1-yl}-1-cyclopropyl-6-fluoro-8-methylquinazoline-2,4(1H,3H)-dione, 5'-D(*AP*CP*CP*AP*AP*GP*GP*T*CP*AP*TP*GP*AP*AP*T)-3', 5'-D(*CP*TP*GP*TP*TP*TP*TP*A*CP*GP*TP*GP*CP*AP*T)-3', ... | | Authors: | Laponogov, I, Pan, X.-S, Veselkov, D.A, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2010-02-16 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Gate-DNA Breakage and Resealing by Type II Topoisomerases
Plos One, 5, 2010
|
|
4BMW
 
 | | Crystal structure of the Streptomyces reticuli HbpS E78D, E81D double mutant | | Descriptor: | EXTRACELLULAR HAEM-BINDING PROTEIN | | Authors: | Wagener, S, Kursula, I, Wedderhoff, I, Groves, M.R, Ortiz de Orue Lucana, D. | | Deposit date: | 2013-05-11 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Iron Binding at Specific Sites within the Octameric Hbps Protects Streptomycetes from Iron-Mediated Oxidative Stress.
Plos One, 8, 2013
|
|
