5DAD
 
 | | Crystal Structure of Human KEAP1 BTB Domain in Complex with Small Molecule TX64014 | | Descriptor: | (6aS,7S,10aS)-8-hydroxy-4-methoxy-2,7,10a-trimethyl-5,6,6a,7,10,10a-hexahydrobenzo[h]quinazoline-9-carbonitrile, Kelch-like ECH-associated protein 1 | | Authors: | Huerta, C, Jiang, X, Trevino, I, Bender, C.F, Swinger, K.K, Stoll, V.S, Ferguson, D.A, Thomas, P.J, Probst, B, Dulubova, I, Visnick, M, Wigley, W.C. | | Deposit date: | 2015-08-19 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Characterization of novel small-molecule NRF2 activators: Structural and biochemical validation of stereospecific KEAP1 binding.
Biochim.Biophys.Acta, 1860, 2016
|
|
8KME
 
 | |
8ODQ
 
 | | SufS-SufU complex from Mycobacterium tuberculosis | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Cysteine desulfurase, NITRATE ION, ... | | Authors: | Elchennawi, I, Carpentier, P, Caux, C, Ponge, M, Ollagnier de Choudens, S. | | Deposit date: | 2023-03-09 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Characterization of Mycobacterium tuberculosis Zinc SufU-SufS Complex.
Biomolecules, 13, 2023
|
|
1UD9
 
 | | Crystal Structure of Proliferating Cell Nuclear Antigen (PCNA) Homolog From Sulfolobus tokodaii | | Descriptor: | DNA polymerase sliding clamp A, ZINC ION | | Authors: | Tanabe, E, Yasutake, Y, Tanaka, Y, Yao, M, Tsumoto, K, Kumagai, I, Tanaka, I. | | Deposit date: | 2003-04-28 | | Release date: | 2004-06-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of Proliferating Cell Nuclear Antigen (PCNA) Homolog From Sulfolobus tokodaii
To be published
|
|
1UP9
 
 | | REDUCED STRUCTURE OF CYTOCHROME C3 FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 AT PH 7.6 | | Descriptor: | CYTOCHROME C3, HEME C, SULFATE ION | | Authors: | Bento, I, Matias, P.M, Baptista, A.M, Da Costa, P.N, Van Dongen, W.M.A.M, Saraiva, L.M, Schneider, T.R, Soares, C.M, Carrondo, M.A. | | Deposit date: | 2003-09-29 | | Release date: | 2004-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Molecular Basis for Redox-Bohr and Cooperative Effects in Cytochrome C3 from Desulfovibrio Desulfuricans Atcc 27774: Crystallographic and Modeling Studies of Oxidized and Reduced High-Resolution Structures at Ph 7.6
Proteins, 54, 2004
|
|
6XQJ
 
 | | Structure of HIV-1 Vpr in complex with the human nucleotide excision repair protein hHR23A | | Descriptor: | Protein Vpr,UV excision repair protein RAD23 homolog A, ZINC ION | | Authors: | Byeon, I.-J.L, Calero, G, Wu, Y, Byeon, C.H, Gronenborn, A.M. | | Deposit date: | 2020-07-09 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of HIV-1 Vpr in complex with the human nucleotide excision repair protein hHR23A.
Nat Commun, 12, 2021
|
|
6ZZC
 
 | | MB_CRS6-1 bound to CrSAS-6_6HR | | Descriptor: | Centriole protein, DODECAETHYLENE GLYCOL, MB_CrS6-1 | | Authors: | Hatzopoulos, G.N, Kukenshoner, T, Banterle, N, Favez, T, Fluckiger, I, Hantschel, O, Gonczy, P. | | Deposit date: | 2020-08-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Tuning SAS-6 architecture with monobodies impairs distinct steps of centriole assembly.
Nat Commun, 12, 2021
|
|
2W36
 
 | | Structures of endonuclease V with DNA reveal initiation of deaminated adenine repair | | Descriptor: | 5'-D(*CP*GP*AP*TP*CP*TP*GP*TP*AP*GP*CP)-3', 5'-D(*GP*CP*BRUP*AP*CP*IP*GP*AP*BRUP*CP*GP)-3', ENDONUCLEASE V | | Authors: | Dalhus, B, Arvai, A.S, Rosnes, I, Olsen, O.E, Backe, P.H, Alseth, I, Gao, H, Cao, W, Tainer, J.A, Bjoras, M. | | Deposit date: | 2008-11-06 | | Release date: | 2009-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Endonuclease V with DNA Reveal Initiation of Deaminated Adenine Repair.
Nat.Struct.Mol.Biol., 16, 2009
|
|
6ZOI
 
 | | A lid blocking mechanism of a cone snail toxin revealed at the atomic level | | Descriptor: | Conknunitzin-C3 mutante | | Authors: | Saikia, C, Altman-Gueta, H, Dym, O, Frolow, F, Gurevitz, M, Gordon, D, Reuveny, E, Karbat, I. | | Deposit date: | 2020-07-07 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | A lid blocking mechanism of a cone snail toxin revealed at the atomic level
To Be Published
|
|
7R36
 
 | | Difference-refined structure of fatty acid photodecarboxylase 2 microsecond following 400-nm laser irradiation of the dark-state determined by SFX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | Authors: | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, De Zitter, E, Schlichting, I, Colletier, J.P, Weik, M. | | Deposit date: | 2022-02-06 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved serial femtosecond crystallography on fatty-acid photodecarboxylase: lessons learned.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7R33
 
 | | Difference-refined structure of fatty acid photodecarboxylase 20 ps following 400-nm laser irradiation of the dark-state determined by SFX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | Authors: | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, De Zitter, E, Schlichting, I, Colletier, J.P, Weik, M. | | Deposit date: | 2022-02-06 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial femtosecond crystallography on fatty-acid photodecarboxylase: lessons learned.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7R34
 
 | | Difference-refined structure of fatty acid photodecarboxylase 900 ps following 400-nm laser irradiation of the dark-state determined by SFX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | Authors: | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, De Zitter, E, Schlichting, I, Colletier, J.P, Weik, M. | | Deposit date: | 2022-02-06 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial femtosecond crystallography on fatty-acid photodecarboxylase: lessons learned.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7R35
 
 | | Difference-refined structure of fatty acid photodecarboxylase 300 ns following 400-nm laser irradiation of the dark-state determined by SFX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | Authors: | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, De Zitter, E, Schlichting, I, Colletier, J.P, Weik, W. | | Deposit date: | 2022-02-06 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial femtosecond crystallography on fatty-acid photodecarboxylase: lessons learned.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6ZRE
 
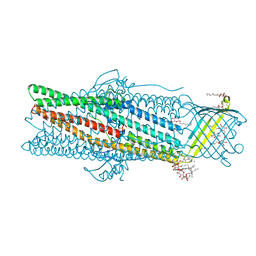 | | Deciphering the role of the channel constrictions in the opening mechanism of MexAB-OprM efflux pump from Pseudomonas aeruginosa | | Descriptor: | Outer membrane protein OprM, PALMITIC ACID, SULFATE ION, ... | | Authors: | Ntsogo Enguene, Y.V, Monlezun, L, Ma, M, Garnier, C, Lascombe, M.B, Salem, M, Guenard, S, Plesiat, P, Llanes, C, Phan, G, Broutin, I. | | Deposit date: | 2020-07-13 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Deciphering the role of OprM constrictions in the opening mechanism of the MexAB-OprM efflux pump from Pseudomonas aeruginosa.
To Be Published
|
|
1HXP
 
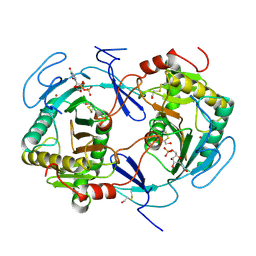 | | NUCLEOTIDE TRANSFERASE | | Descriptor: | BETA-MERCAPTOETHANOL, FE (III) ION, HEXOSE-1-PHOSPHATE URIDYLYLTRANSFERASE, ... | | Authors: | Wedekind, J.E, Frey, P.A, Rayment, I. | | Deposit date: | 1995-06-09 | | Release date: | 1996-11-08 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of galactose-1-phosphate uridylyltransferase from Escherichia coli at 1.8 A resolution.
Biochemistry, 34, 1995
|
|
7RKL
 
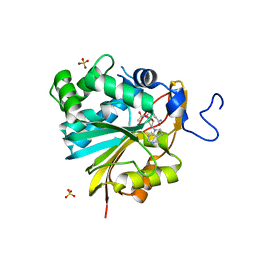 | | Structure of Nicotinamide N-Methyltransferase (NNMT) in complex with II399 (P1 space group) | | Descriptor: | 3-[3-(acetyl{[(1R,2R,3S,4R)-4-(4-chloro-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-2,3-dihydroxycyclopentyl]methyl}amino)prop-1-yn-1-yl]benzamide, NNMT protein, SULFATE ION | | Authors: | Yadav, R, Noinaj, N, Iyamu, I.D, Huang, R. | | Deposit date: | 2021-07-22 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Exploring Unconventional SAM Analogues To Build Cell-Potent Bisubstrate Inhibitors for Nicotinamide N-Methyltransferase.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
5OGW
 
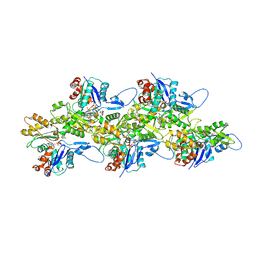 | | Cryo-EM structure of jasplakinolide-stabilized malaria parasite F-actin at near-atomic resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-1, Jasplakinolide, ... | | Authors: | Pospich, S, Kumpula, E.-P, von der Ecken, J, Vahokoski, J, Kursula, I, Raunser, S. | | Deposit date: | 2017-07-13 | | Release date: | 2017-09-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Near-atomic structure of jasplakinolide-stabilized malaria parasite F-actin reveals the structural basis of filament instability.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7RKB
 
 | | Crystal Structure of Putative Pterin Binding Protein (PruR) from Klebsiella pneumoniae in Complex with Neopterin | | Descriptor: | CHLORIDE ION, L-NEOPTERIN, Pterin Binding Protein, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-22 | | Release date: | 2022-08-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Putative Pterin Binding Protein (PruR) from Klebsiella pneumoniae in Complex with Neopterin.
To Be Published
|
|
5DAF
 
 | | Crystal Structure of Human KEAP1 BTB Domain in Complex with Small Molecule TX64063 | | Descriptor: | (5aS,6S,9aS)-7-hydroxy-2,6,9a-trimethyl-3-(pyridin-3-yl)-4,5,5a,6,9,9a-hexahydro-2H-benzo[g]indazole-8-carbonitrile, Kelch-like ECH-associated protein 1 | | Authors: | Huerta, C, Jiang, X, Trevino, I, Bender, C.F, Swinger, K.K, Stoll, V.S, Ferguson, D.A, Thomas, P.J, Probst, B, Dulubova, I, Visnick, M, Wigley, W.C. | | Deposit date: | 2015-08-19 | | Release date: | 2016-08-10 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Characterization of novel small-molecule NRF2 activators: Structural and biochemical validation of stereospecific KEAP1 binding.
Biochim.Biophys.Acta, 1860, 2016
|
|
2YM5
 
 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | (3-{4-[(2S)-2-(AMINOMETHYL)MORPHOLIN-4-YL]-7H-PYRROLO[2,3-D]PYRIMIDIN-5-YL}PHENYL)METHANOL, 1,2-ETHANEDIOL, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Reader, J.C, Matthews, T.P, Klair, S, Cheung, K.M.J, Scanlon, J, Proisy, N, Addison, G, Ellard, J, Piton, N, Taylor, S, Cherry, M, Fisher, M, Boxall, K, Burns, S, Walton, M.I, Westwood, I.M, Hayes, A, Eve, P, Valenti, M, Brandon, A.H, Box, G, vanMontfort, R.L.M, Williams, D.H, Aherne, G.W, Raynaud, F.I, Eccles, S.A, Garrett, M.D, Collins, I. | | Deposit date: | 2011-06-06 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure-Guided Evolution of Potent and Selective Chk1 Inhibitors Through Scaffold Morphing.
J.Med.Chem., 54, 2011
|
|
8OJ2
 
 | |
6YZZ
 
 | | Arabidopsis thaliana Naa50 in complex with AcCoA | | Descriptor: | ACETYL COENZYME *A, N-alpha-acetyltransferase 50 | | Authors: | Weidenhausen, J, Kopp, J, Lapouge, K, Sinning, I. | | Deposit date: | 2020-05-07 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural and functional characterization of the N-terminal acetyltransferase Naa50.
Structure, 29, 2021
|
|
2YM6
 
 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 1-[(2R)-4-(9H-pyrido[4',3':4,5]pyrrolo[2,3-d]pyrimidin-4-yl)morpholin-2-yl]methanamine, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Reader, J.C, Matthews, T.P, Klair, S, Cheung, K.M.J, Scanlon, J, Proisy, N, Addison, G, Ellard, J, Piton, N, Taylor, S, Cherry, M, Fisher, M, Boxall, K, Burns, S, Walton, M.I, Westwood, I.M, Hayes, A, Eve, P, Valenti, M, Brandon, A.H, Box, G, vanMontfort, R.L.M, Williams, D.H, Aherne, G.W, Raynaud, F.I, Eccles, S.A, Garrett, M.D, Collins, I. | | Deposit date: | 2011-06-06 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-Guided Evolution of Potent and Selective Chk1 Inhibitors Through Scaffold Morphing.
J.Med.Chem., 54, 2011
|
|
8OJ1
 
 | |
6Y1L
 
 | | Crystal structure of the paraoxon-modified A.17 antibody FAB fragment - L47R mutant | | Descriptor: | DIETHYL PHOSPHONATE, FAB A.17 L47R mutant HEAVY CHAIN, FAB A.17 L47R mutant Light CHAIN, ... | | Authors: | Chatziefthimiou, S, Mokrushina, Y, Smirnov, I, Gabibov, A, Wilmanns, M. | | Deposit date: | 2020-02-12 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
